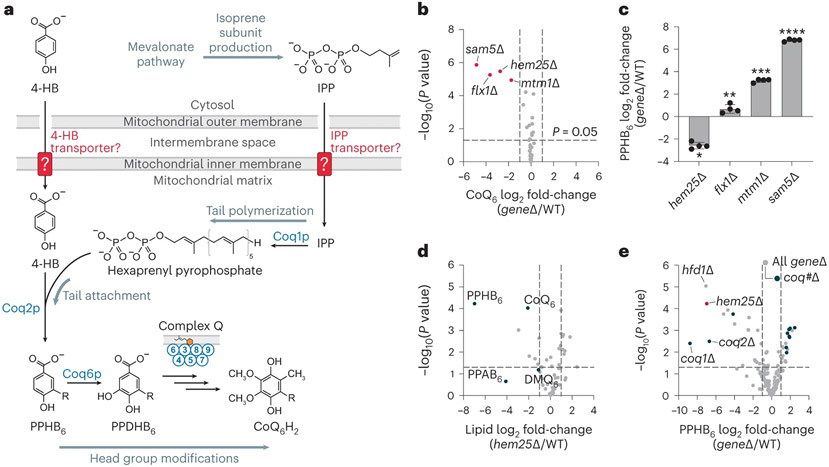
Fig. 1: A targeted genetic screen identifies Hem25p as a potential transporter of CoQ precursors.
a, Schematic of CoQ biosynthesis in S. cerevisiae. 4-HB, 4-hydroxybenzoate; IPP, isopentenyl pyrophosphate; PPHB6, polyprenyl-hydroxybenzoate; PPDHB6, polyprenyl-dihydroxybenzoate. b, Relative CoQ abundance in all geneΔ strains compared to wild type (WT) versus statistical significance. Hits with significantly decreased levels of CoQ are highlighted (mean, n = 3 biologically independent samples, two-sided Student’s t-test). c, Relative PPHB levels in each of the hits from b (*P = 3.1 × 10−6 WT versus hem25Δ, **P = 0.0139 WT versus flx1Δ, ***P = 4.1 × 10−8 WT versus mtm1Δ, ****P = 1.3 × 10−10 WT versus sam5Δ; mean ± s.d., n = 4 biologically independent samples, two-sided Student’s t-test). d, Relative lipid abundances in hem25Δ yeast compared to WT versus statistical significance with CoQ and CoQ biosynthetic intermediates highlighted. PPAB6, polyprenyl-aminobenzoate; DMQ6, demethoxy-coenzyme Q. e, Relative PPHB abundances versus statistical significance across all Y3K geneΔ strains, with hem25Δ, hfd1Δ and coq#Δ strains highlighted. For d and e, raw data from the Y3K dataset33 (respiration-RDR (respiration deficiency response) condition) are displayed as the mean from three biologically independent samples, with two-sided Student’s t-test used for both panels. Numerical data are available as Source data.