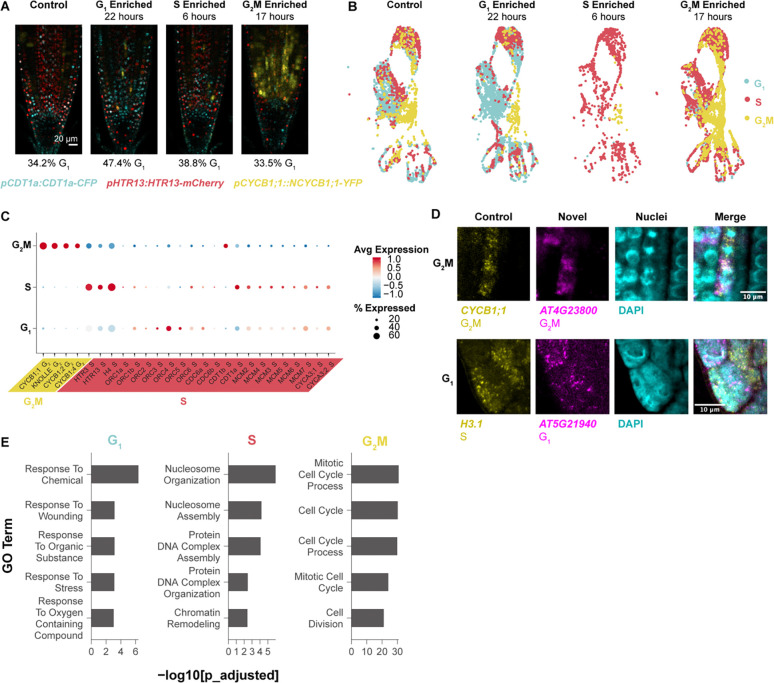
Figure 1: Single-cell phase synchronized cells yield robust transcriptional markers for each phase of the cell cycle.
(A) Representative images of phase enrichments achieved with HU synchronization using seedlings expressing the PlaCCI reporter. The G1 phase reporter pCDT1a:CDT1a-CFP is shown in cyan, the S phase reporter pHTR13:HTR13-mCherry is shown in red, and the G2/M phase reporter pCYCB1;1:NCYCB1;1-YFP is shown in yellow. The percentage of cells in G1 (CFP positive cells/All cells), determined from 3D segmentation of z stacks, is shown below each panel. The hours noted above each panel refer to the treatment time each batch of roots was exposed to 10mM HU for. (B) An unsynchronized control and three single cell profiles collected at specific times after synchronization integrated in UMAP with cells color coded by phase determination. Cells from each time point were separated after integration. (C) A dot plot showing expression of gold standard cell-cycle phase markers, showing known G2/M-phase markers (CYCBs) followed by known S-phase markers. (D) In situ hybridization of novel G1 and G2/M probes. Known markers are shown in yellow and new markers in magenta. The new G2/M marker is hybridized with a known G2/M marker, showing overlap. The new G1 marker is hybridized with a known S marker, showing spatial anticorrelation. (E) The top five most statistically significantly enriched GO terms among the top 50 phase marker set for each phase. See also Figures S1–S5 and Tables S1–S3.