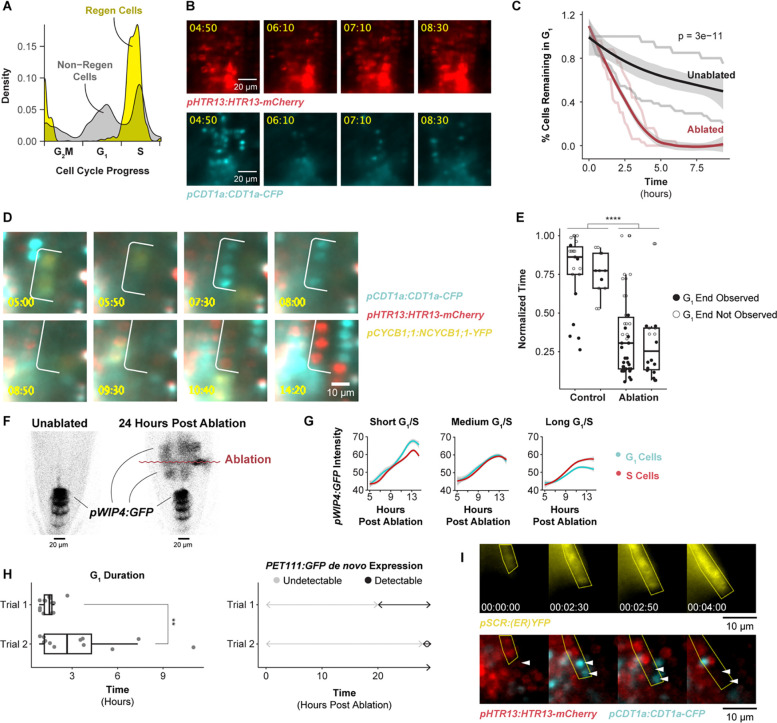
Figure 3: The G1 phase of the cell cycle is dramatically truncated in regenerating cells.
(A) Summary of the frequency of a given cell cycle phase in regenerating (yellow) and non-regenerating (grey) cells. Cells are aligned along a cell cycle pseudotime on the x axis, with their density shown on the y axis with G1 predominant in non-regenerating cells and almost absent in regenerating cells. (B) Representative images of cells coordinately exiting G1 following tissue damage (bottom). S-phase cells (top) serve as a control showing a continuous strong signal (no depletion) in the same roots. Time is shown in hours:minutes and 00:00 is the time of the ablation. (C) Quantification of the coordinated G1 exit shown as a survival analysis of the population of G1 cells identified at the beginning of the time lapse. The y axis represents the fraction of G1 cells from time zero (the first frame of the time lapse) still in G1 at time n. The x axis shows time since the beginning of the time lapse. Time zero for each ablated root is 3 hpa. Two unablated and two ablated roots were analyzed. (n = 165 cells, p-value = 3e-11, log rank test). Lightly colored lines are individual replicates, and the bold line is a LOESS regression of the two trials. P-value of survival rates is significantly different between ablated and unablated roots using the log rank test. (D) Representative time-lapse series of a short G1 in an ablated root in which cells pass through the phase in as little as 2 hr. Time is shown in hours:minutes and 00:00 is the time of the ablation. (E) Quantification of G1 duration in control and ablated roots for two trials. Time is normalized within each root to the duration of the time lapse in which it was measured, where the y-axis represents the time in which a cell was in G1 divided by the duration of the time lapse. Filled dots represent cells in which the end of G1 was observed. Each dot represents an individual cell. (n=94 cells, p-value = 3.221e-09, Mann-Whitney U Test). (F) Representative image of the pWIP4:GFP expression domain before ablation and 24 hpa. The red wavy line marks the location of the ablation. (G) Quantification of pWIP4:GFP signal over time in G1 and S phase cells, with different plots showing analysis of cells grouped by the length of G1 or S. Short = 3 hr, medium = 6 hr, long = 9 hr. (n = 650 cells in 1 root). (H) Quantification of G1 duration in two roots (left) and the timing of PET111 expression establishment in the regeneration zone in the same two roots (right) showing the association between G1 duration and PET11 appearance. Each dot represents an individual cell. Trials refer to individual root time lapses (p-value = 0.00413 Mann-Whitney test). (I) Representative image of the expansion of the expression domain of the pSCR:erYFP reporter during regeneration. The SCR expression domain is outlined with a yellow ROI on both the upper and lower panels. Panels were chosen to show the cell marked with an arrow just before division, while in G1, and after each daughter exits G1. The first frame of this montage is 26 hpa and the time stamp of each frame are as follows (day:hr:min): 00:00:00, 00:02:30, 00:02:50, 00:04:00. See also Figure S7, Table S4–S5s, and Movies S2–S3.