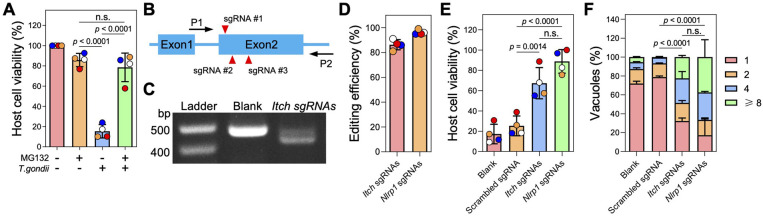
Fig. 3. Knockout of Itch impairs Toxoplasma-induced NLRP1-mediated cell death of Lewis rat BMDMs.
A. Lewis rat BMDMs pre-treated with 0.5 μM proteasome inhibitor MG132 or left untreated for 2 h were infected with Toxoplasma parasites (MOI = 1) for 24 h. Macrophage viability was measured via MTS assay. Data are displayed as mean ± SD with independent experiments (n = 4) indicated by the same color dots. Significance was determined with one-way ANOVA with Tukey’s multiple comparisons test.
B. Schematic illustration of sgRNA targeting sites in the first two exons of rat ITCH locus. P1 and P2 are the primers used for amplifying the sgRNA targeting region and verification by Sanger sequencing.
C. PCR amplification of the Itch sgRNA targeting region using P1 and P2 from Lewis rat BMDMs transfected with Cas9 protein only (blank) or Cas9 protein assembled with Itch sgRNAs.
D. Lewis rat BMDMs were transfected with in vitro assembled CRISPR/Cas9 ribonucleoprotein containing indicated sgRNAs. The editing efficiency of Itch or Nlrp1 was analyzed from the Sanger sequencing products (shown in c) using the Inference of CRISPR Edits (ICE) online tool (https://www.synthego.com/products/bioinformatics/crispr-analysis). The percentage of CRISPR edited Itch or Nlrp1 is displayed as mean ± SD with independent experiments (n = 4) indicated by the same color dots.
E. Lewis rat BMDMs generated in (d) were infected with wild-type Toxoplasma (MOI = 1) for 24 h. Macrophage viability was measured via MTS assay. Data are displayed as mean ± SD with independent experiments (n = 4) indicated by the same color dots. Significance was determined with one-way ANOVA with Tukey’s multiple comparisons test.
F. Lewis rat BMDMs transfected with in vitro assembled CRISPR/Cas9 ribonucleoprotein containing indicated sgRNAs were infected with wild-type Toxoplasma (MOI = 0.5) for 24 h. The number of parasites per vacuole was quantified by microscopy. A total of 100 to 120 vacuoles were counted per experiment. Data are displayed as mean + SD with 2 independent experiments. Significance was determined with two-way ANOVA with Tukey’s multiple comparisons test.