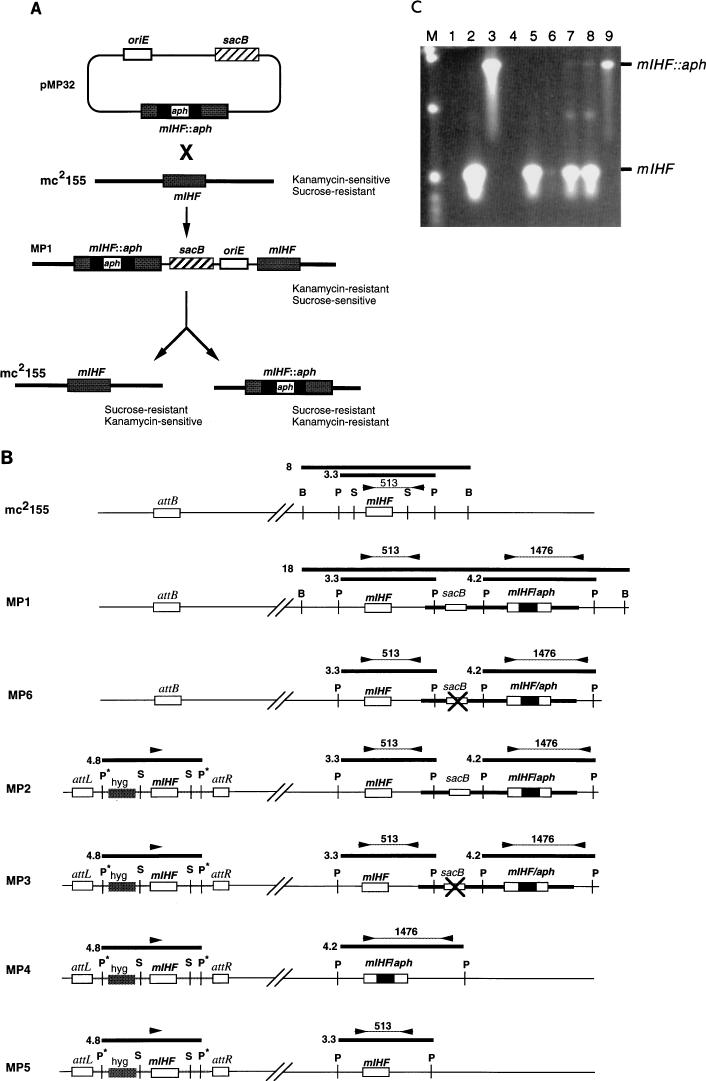
FIG. 3.
Allelic replacement of mIHF. (A) Strategy for allelic replacement of M. smegmatis mIHF. Transformation of M. smegmatis mc2155 with plasmid pMP32 generates a strain (MP1) containing an integrated copy of the plasmid at the mIHF locus. Strain MP1 is resistant to kanamycin but sensitive to sucrose due to the presence of the sacB gene. Selection of sucrose-resistant colonies can give rise to two alternative products depending on where recombination occurs. If the recombination event is on the same side of aph as the initial integration event, then the original strain (mc2155) is regenerated; if it is on the other side, then the mIHF gene is replaced by an interrupted copy. (B) Schematic representation of strains used or generated in allelic replacement experiments. At the top, the mIHF locus of M. smegmatis mc2155 is represented with the positions of restriction sites for BamHI (B), PstI (P), and SalI (S) indicated; the unlinked attB site is also shown. Also shown are relevant parts of the chromosomes of strains MP1, MP6, MP2, MP3, MP4, and MP5. Strain MP1 is a derivative of mc2155 created by insertion of plasmid pMP32 by homologous recombination at the mIHF locus; MP6 is a derivative of MP1 that is similar to MP1 as determined by Southern hybridization and PCR analyses but is sucrose resistant and probably contains an inactivating mutation within the sacB gene (×). Strain MP2 was derived from MP1 by transformation with an integration-proficient plasmid containing the mIHF gene that integrates site specifically at the attB locus. Strains MP3, MP4, and MP5 are sucrose-resistant derivatives of MP2 that have an inactivating mutation in sacB (MP3) or have undergone recombination at the mIHF locus to leave only the wild-type mIHF gene (MP5) or a replacement by an interrupted mIHF gene (MP4). DNA fragments generated by restriction enzyme digests that hybridize with an mIHF-specific DNA probe are shown as thick horizontal lines with their sizes in kilobases. The positions of primers used in PCR characterization experiments are shown as arrowheads, and the sizes of PCR products are shown in base pairs. Restriction sites originating from plasmid vector sequences are shown with an asterisk. (C) PCR amplification of the mIHF locus in M. smegmatis strains. DNAs from various M. smegmatis strains were used for PCR amplication with the primers shown in panel B, and the products were separated by agarose gel electrophoresis. DNAs used were from pMP18 (containing the wild-type mIHF gene) (lane 2), pMP32 (containing the aph-interrupted mIHF gene) (lane 3), pMP28 (which contains wild-type mIHF but lacks one of the primer binding sites) (lane 4), mc2155 (lane 5), MP1 (lane 6), MP2 (lane 7), MP3 (lane 8), and MP4 (lane 9). Lane 1 contains no DNA. The positions of the 513-bp fragment amplified from the wild-type mIHF gene and the 1,476-bp fragment from the aph-interrupted gene are indicated. Note that when both the wild-type and interrupted mIHF loci are present in the same strain (i.e., MP1, MP2, and MP3) the smaller product is preferentially amplified. Plasmid pMP32 was constructed by insertion of a sacB fragment into pMP27, a pUC119 derivative that contains the mIHF gene interrupted by the aph gene at the EcoNI site.