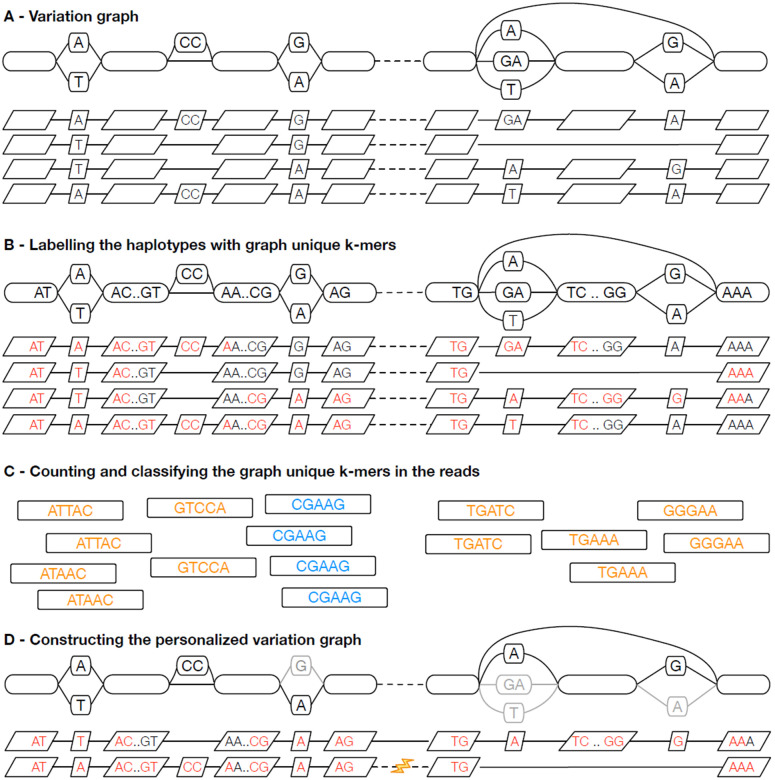
Figure 1: Illustrating haplotype sampling at adjacent blocks in the pangenome.
(A) A variation graph representing adjacent locations in the pangenome, composed of a bidirected sequence graph (top) and a set of embedded haplotypes (below); the dotted lines represent the boundary between the two blocks. (B) k-mers that occur once within the graph, termed graph-unique k-mers, are identified in the haplotypes; here k = 5 and graph-unique k-mers are colored red. The presence and absence of these graph-unique k-mers identifies each haplotype. (C) The graph-unique k-mers are counted in the reads, and based upon counts classified as present, likely heterozygous (shown in orange), present, likely homozygous (shown in blue), or absent (all red kmers in (B) not identified in the reads). (D) Using the identified graph-unique k-mer classifications, a subset of haplotypes are selected at each location, defining a personalized pangenome reference subgraph of the larger graph. Where needed, recombinations are introduced (see lightning bolt) to create contiguous haplotypes.