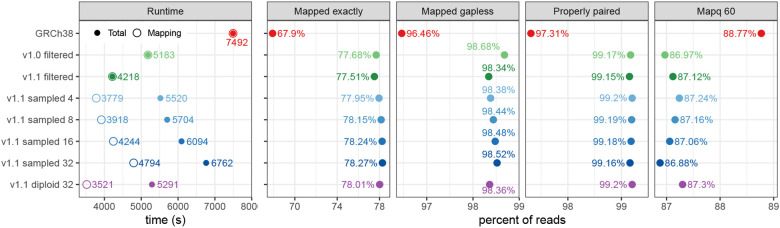
Figure 2: Mapping 30x NovaSeq reads for HG002 to GRCh38 (with BWA-MEM) and to HPRC graphs (with Giraffe).
The graphs (y-axis) are Minigraph–Cactus graphs built using GRCh38 as the reference. For the sampled graphs, we tested sampling 4, 8, 16, and 32 haplotypes. For the v1.1 diploid graph, 32 candidate haplotypes were used for diploid sampling. We show the overall running time and the time spent for mapping only (left), and the fraction of reads with an exact, gapless, properly paired, and mapping quality 60 alignment.