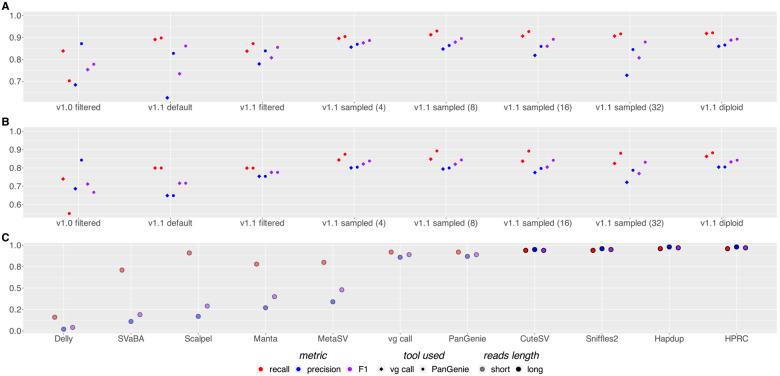
Figure 4: SVs benchmark evaluation.
(A) Precision, recall, and F1 scores of both vg call and PanGenie for different pangenome reference graphs on the GIAB v0.6 Tier1 call set. Graphs were built using GRCh38 as the reference. (B) As with (A) but using a benchmark set of SVs created with DipCall from the T2T v0.9 HG002 genome assembly, comparing genome-wide but excluding centromeres. (C) Comparing the performance of PanGenie and vg call using the 1.1 diploid graph to other genotyping methods. Illumina short reads were used with Delly [27], SVaBA [37], Scalpel [8], Manta [4] and MetaSV [22] as well as with vg call [12] and PanGenie [6]. Also shown are long-read methods (CuteSV [15], Sniffles2 [32], Hapdup [17], and Human Pangenome Reference Consortium de novo assemblies [20]).