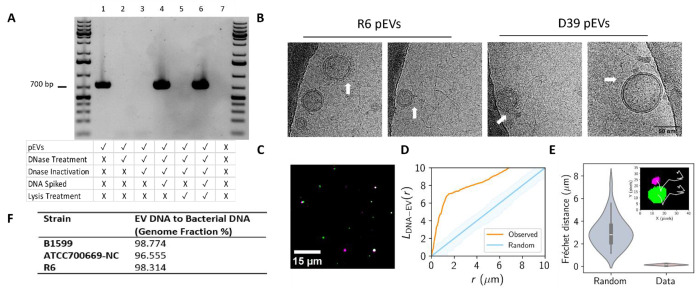
Fig.1: Genomic DNA is present on the surface of pEVs.
(A) PCR analysis of pEV DNA localization. pEVs underwent a series of treatments to determine the location of DNA (representative gel, n=3). At each treatment step, including the untreated pEVs, a sample was saved for PCR amplification and subsequent gel electrophoresis. The rows below each lane indicate each sample’s treatment. DNase treatment was performed with 1 U Turbo DNase at 37 °C for 30 minutes. DNase inactivation was accomplished with 5 μM EDTA (f.c.) for 10 minutes at 75 °C. Samples that were spiked with DNA had 50 ng of genomic DNA added. Lysis treatment was performed with 1% triton (f.c.) at 65 °C for 10 minutes The negative control in the last lane includes only 1x PBS, the buffer used for PEV elution from SEC. Primers targeted the gene gapdh. First and last lanes include the GeneRuler 1 kb Plus DNA Ladder (Invitrogen). (B) Images selected from cryo-electron micrographs of pEVs from R6 and D39 that display nucleic acid strands surrounding pEVs. White arrows indicate point of association between pEVs and nucleic acid strands. All images are the same scale (scale bar in final image). (C-D) Spatial analysis of DNA and pEV co-localization. (C) Representative image of DNA particles, false colored in green, and pEV particles, false colored in magenta. pEV sample was treated with PicoGreen and DiD to label DNA and the pEV membrane, respectively. Scale bar is indicated. (D) Ripley’s cross-L function for clustering of simulated random (blue) and observed (orange) pEV particles to DNA particles as a function radius, r. Data represent n=4 independent fields of view of DNA and pEV molecules (n=175 pEV particles and n=477 DNA particles). Shading represents the acceptance region for a hypothesis test of complete spatial randomness, with significance level 5%, using the envelopes of the L-functions of 1000 simulations. (E) DNA and pEV co-diffusion. Frechet distances computed for simulated (Random) and experimental (Data) of pEV-DNA particle trajectories. Data represent n=4 pEV-DNA pairs. Random Fréchet distance determined using diffusion coefficients for each pair determined in SFig 2. Inset: representative trajectories (white) from a DNA (green) and pEV (magenta) pair. (F) Sequencing of DNA isolated from pEVs and their parent bacterium from three unencapsulated pneumococcal strains (n=1 per strain).