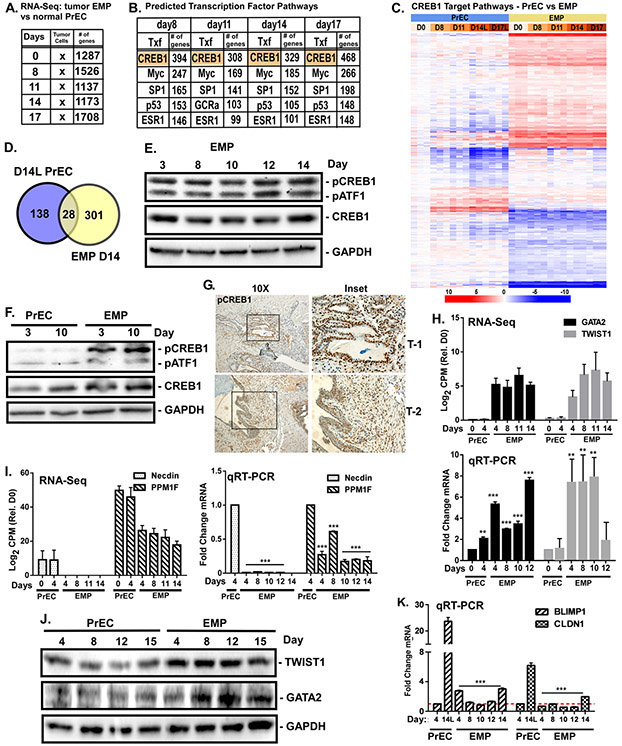
Figure 5. CREB1 targets different genes upon oncogenic transformation.
EMP cells (PrEC overexpressing ERG (E), MYC (M), and shPTEN (P)) subjected to the differentiation protocol for 0-17 days. RNA sequencing was performed on biological triplicates in parallel with PrECs (Fig.1). Differentially expressed genes with p<0.05 at each time point relative to normal basal cells were identified. A) Metacore analysis of transcription factors (Tfx) likely involved in regulating differentially expressed genes. B) Top 5 Tfx shown for each time point and the number of genes regulated by that Tfx are indicated. C) Heat map comparing CREB1-regulated genes in PrECs versus EMP cells. D) Comparison of CREB1-regulated genes from Day 14 luminal (L) PrECs vs. EMP cells. E,F) Levels of pCREB1 and pATF1 (Ser133), CREB1, and ATF1 measured by immunoblotting during differentiation in E) EMP or F) EMP versus PrEC. GAPDH served as loading control. G) IHC staining of pCREB1 in two EMP orthotopic xenograft tumors. H,I) Transcript counts from RNA-seq (log2 CPM) and qRT-PCR mRNA levels (log2 fold change) of EMP CREB1-specific target genes: H) GATA2 and Twist1, and I) NDN (Necdin) and PPM1F. J) Levels of Twist1 and GATA2 in differentiating PrEC versus EMP measured by immunoblotting. K) Levels of PMDR1 (BLIMP1) and CLDN1 mRNA measured by qRT-PCR from differentiating PrEC vs EMP. **p<0.01; ***p<0.001; n=4; error bars SD relative to PrEC.