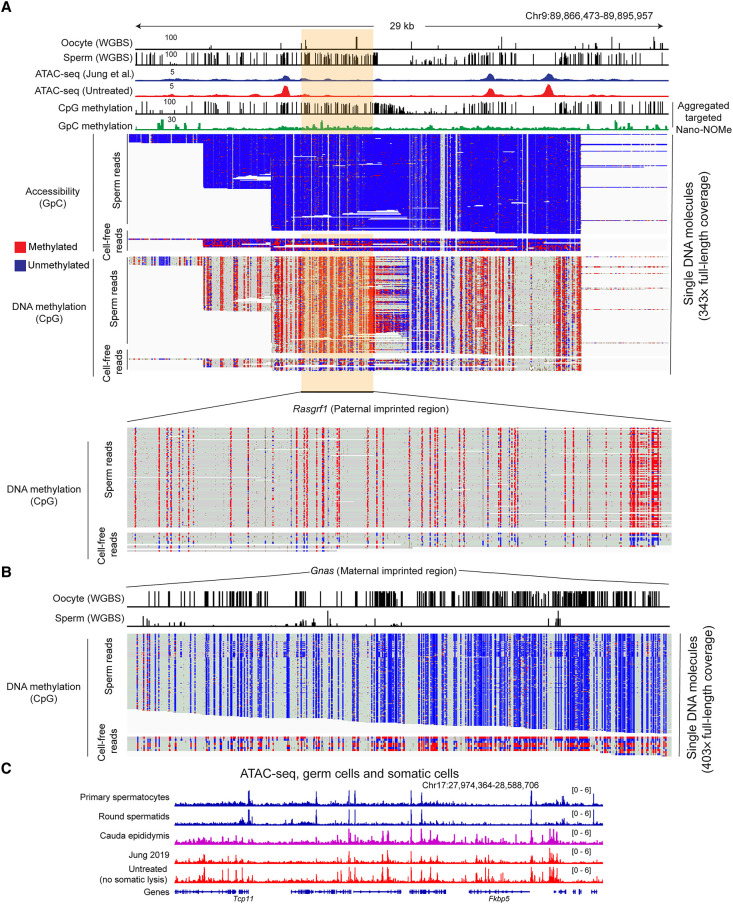
Figure 4.
Contaminating cell-free DNA is of somatic origin. (A,B) Nano-NOMe-seq data for two imprinting control regions in untreated sperm. Untreated sperm were subject to M.CviPI-driven GpC methylation; then genomic DNA was extracted after DTT treatment to obtain DNA from both contaminating material as well as sperm; and resulting DNA was subject to long-read sequencing by Oxford Nanopore. Resulting methylation calls (red and blue represent methylated and unmethylated cytosine, respectively) are shown separately for CpG methylation and GpC methylation (analysis was restricted to HCG and GCH, where H represents A/C/T, to avoid ambiguous GCG methylation), as indicated. In addition, reads are separated based on the extent of GpC methylation. The majority of reads show low (5.5 ± 3.4%) GpC methylation, whereas a small fraction of reads show >20% GpC methylation and represent accessible DNA molecules in untreated sperm preparations. Importantly, DNA molecules protected from M.CviPI, arising from the bona fide sperm genome, show the 0% or 100% methylation expected at imprinting control regions in germline samples. In contrast, the small fraction of GC-methylated reads, reflecting accessible cell-free chromatin, includes a mix of methylated and unmethylated ICRs consistent with a somatic origin for these DNA molecules. (C) ATAC-seq data for the indicated samples, including ATAC-seq for cauda epididymal epithelium. Key here is the strong agreement between ATAC profiles for untreated sperm (representing contaminating chromatin) and cauda epididymal epithelium (see also Supplemental Fig. S2B).