ABSTRACT
The human gut microbiota constitutes a vast and complex community of microorganisms. The myriad of microorganisms present in the intestinal tract exhibits highly intricate interactions, which play a crucial role in maintaining the stability and balance of the gut microbial ecosystem. These interactions, in turn, influence the overall health of the host. The mammalian gut microbes have evolved a wide range of mechanisms to suppress or even eliminate their competitors for nutrients and space. Simultaneously, extensive cooperative interactions exist among different microbes to optimize resource utilization and enhance their own fitness. This review will focus on the competitive mechanisms among members of the gut microorganisms and discuss key modes of actions, including bacterial secretion systems, bacteriocins, membrane vesicles (MVs) etc. Additionally, we will summarize the current knowledge of the often-overlooked positive interactions within the gut microbiota, and showcase representative machineries. This information will serve as a reference for better understanding the complex interactions occurring within the mammalian gut environment. Understanding the interaction dynamics of competition and cooperation within the gut microbiota is crucial to unraveling the ecology of the mammalian gut microbial communities. Targeted interventions aimed at modulating these interactions may offer potential therapeutic strategies for disease conditions.
KEYWORDS: Microbiota, positive interaction, negative interaction, mechanisms, gut ecosystem
Introduction
The mammalian gastrointestinal tract (GI) is a complex ecosystem composed of a vast community of microbes known as the gut microbiota, which plays a pivotal role in host health.1 The human gut microbiota comprises bacteria, archaea, eukaryotes and viruses. It has co-evolved with the host for millions of years.2–4 Over 100 trillion microorganisms, representing more than 1000 different species, thrive within the human intestinal tract, and the composition can vary significantly between individuals according to different factors including diet, host genetics and age etc.5,6 Imbalances in the composition of these intestinal microbes have been linked to an increasing number of host diseases and syndromes.1
It is generally recognized that the neonate’s microbial colonization event first occurs at birth through the birth canal and subsequently from the immediate environment.7–9 This colonization then continually matures and stabilizes during the first 3 years of life. In adults, the composition of the gut microbiota remains relatively stable, except in cases of extreme external stressors, such as antibiotics, diet, and infection.10 The different species hosted within the human gut form a complex ecological interaction web, interacting with one another either positively or negatively to create and adapt to suitable living conditions.
Competition and cooperation between microbes play a crucial role in shaping community composition and functioning in the gut.11 Within these communities, bacteria compete with their neighbors for nutrients, including carbon, nitrogen, metals, phosphate, as well as for space where these nutrients are more abundant. Microbial communities also engage in cooperation to aid in resource digestion, combat antibiotics, or manage other environmental stresses.12 Previous studies have indicated that there is often more pronounced competition between bacterial species than cooperation,13–15 yet an increasing number of recent studies highlight the prevalence and ecological significance of cooperative phenotypes in the mammalian gut.16
There are trillions of microorganisms inhabiting the intestinal tract, and the microbiota members may engage in direct or indirect interactions. Understanding the interactions among these gut bacteria provides insight into the forces shaping the ecology of the human gut microbiota. This review highlights the diverse competitive and cooperative interactions among the dominant gut flora, aiding in the comprehension of the dynamics of the gut ecosystem. Importantly, by embracing this dynamic perspective, we can better decipher the underlying mechanisms that shape community composition and function in both health and disease.
Competitive interactions in the human gut
The microorganisms inhabiting the human GI tract live in close contact with each other in a complex and dynamic relationship (Figure 1). Exposure to various environmental factors can strongly influence microbial interactions, resulting in positive, negative, or neutral outcomes. A strain is considered to be competitive if it shows a phenotype that decreases the survival or reproduction of others.14 It has long been recognized that there are two main types of competition: exploitative competition and interference competition, both of which are categorized as real competition17. Interference competition is a direct form of competition in which one individual actively harms another or engages in chemical warfare, while exploitative competition occurs indirectly through competition for common resources17. Interference-based competition usually antagonizes competitors by releasing bacteriocins directly or in a secretion system-dependent manner.
Figure 1.
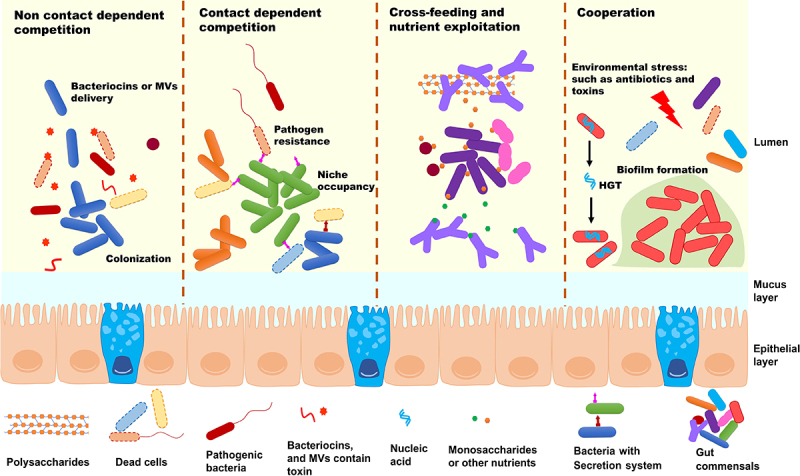
Scheme of gut microbial interaction in gut. Bacterial interactions, whether competitive or cooperative, involve a significant investment of energy and stringent regulatory control. Competitive interactions can take the form of exploitation, or interference, while positive interaction mostly related to nutrients cross-feeding and adaptation to environmental stress. Secretion of toxin by contact-dependent or independent manner will confers the bacterial colonization, facilitate niche occupancy, and also eliminate the pathogenic bacteria. Positive interactions among bacteria primarily revolve around optimizing resource utilization and adapting to environmental stresses such as antibiotics or toxins, which will enhance bacterial fitness to complex gut environment.
Bacterial secretion systems are a class of protein complexes that translocate cargos across the cytoplasmic membrane.18–21 To date, there are 11 known bacterial protein secretion systems with considerable evolutionary and structural diversity. Most of the secretion systems are specific to Gram-negative bacteria, while only a few have been identified in Gram-positive bacteria. In certain cases, bacterial pathogens utilize these dedicated secretion systems to secrete effectors into the environment or directly into the cytoplasm of the target cells for pathogenicity or survival, or for promoting inter-bacterial competition.18,22,23 Among the 11 identified bacterial secretion systems, the Type I secretion system (T1SS), Type IV secretion system (T4SS), Type V secretion system subtype b (T5SS b), Type VI secretion system (T6SS), and Type VII secretion system (T7SS) have been proven to deliver antibacterial toxins.18,24,25 Among these, T6SS and T7SS are prevalent in gut symbionts. In the following sections, we will provide an overview of the primary types of interference interactions mediated by secretion systems and bacteriocins that are commonly observed in the human gut ecosystem.
Interference competition
Type VI secretion system
Over the past years, T6SS has received considerable attention due to its contact-dependent cell-cell interactions with both bacteria and eukaryotic hosts.26 T6SS is widely distributed in Gram-negative bacteria.27 In human microbial ecosystems, T6SS loci have been found both in Bacteroidetes and Proteobacteria. The T6SS is a contractile nanomachine that injects effector proteins directly from the bacterial cytoplasm into target cells to exert their action28(Figure 2). Scientists, through mathematical modeling, predicted that there are over one billion T6SS effector transmission events per minute per gram of colonic contents of gnotobiotic mice, indicating a critical potential of T6SS in shaping the gut ecosystem29. Over the past decade, significant progress has been made in elucidating the structure of the T6SS and understanding how it confers a competitive advantage to bacterial communities. In silico analysis revealed that T6SS clusters are prevalent in human gut Bacteroidales species with three distinct genetic architectures, GA1, GA2, and GA3.30 Of the three GAs, GA1 and GA2 T6SS gene clusters are encoded on integrative conjugative elements and transferred extensively among numerous species of Bacteroidales, whereas GA3 T6SSs are only found in B. fragilis.30–32
Figure 2.
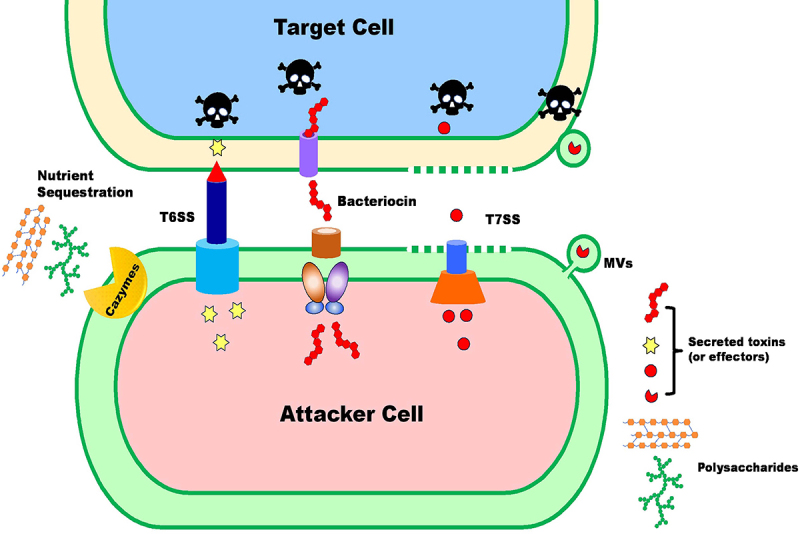
Main competition machineries in gut microbes. Bacterial competition in the gut can be mediated by contact-dependent secretion systems such as T6SS or T7SS, which lyse neighboring cells by injecting toxic effectors into host cells or the environment. Additionally, bacteria can harm distant cells by releasing bacteriocins directly into the environment or by carrying them via MVs. Moreover, exploiting nutrients in the environment through the direct acquisition or release of enzymes is also an important mode of competition among intestinal bacteria.
A systematic gut microbiota metagenomic analysis revealed that GA3 T6SS contributes to early life competition and colonization, and may finally influence the gut microbiota composition in the human gut.33 In vitro work demonstrates that, GA3 T6SS antagonizes almost all species of gut Bacteroidales, exhibiting a more strain-specific impact in a mouse gut model.29,34 Furthermore, the GA3 T6SS mediated killing rate of gut commensals observed was much lower than the Lotka – Volterra model has predicted, suggesting complex and dynamic interactions of microbes in the gut ecosystem.29 In the mouse gut, the abundance of T6SS producers will increase while competing with sensitive competitors and significantly decrease if co-existent with resistant or non-interactive strangers.35 These findings suggest that GA3 T6SS-dependent competition mechanism within a microbial community is intricate and heavily relies on the surrounding microbial lineages inhabiting the same ecological niches, as well as their sensitivity to T6SS antagonistic actions (Figure 1). A genomic and metagenomic data based study indicated that GA2 and GA1 T6SS are widespread in Bacteroidales isolated from gut but absent in bacteria isolated from oral cavity or vagina, suggesting fitness advantage conferred by T6SS in gut environment36. In our recent work, we identified a variety of toxic effectors in GA2 variable regions and experimentally confirmed the periplasmic toxicity.37 However, strong antagonistic effect (1–3 log killing) has not been observed for either the GA1 or GA2 T6SSs. Currently, the main function of the GA1 and GA2 T6SS gene clusters remains to be determined. A very interesting study of Madeline et al. identified that the integration of the ICE encoding GA1 into GA3-containing B. fragilis genome will deactivate its GA3-dependent antagonism. Notably, instead, it will enhance the fitness of B. fragilis by GA1 T6SS dependent antagonism in the mouse gut.38 Extensive intra-ecosystem transferring of GA1 ICE may equip the GA3-containing B. fragilis with additional weapon, which will prevent them from targeting Bacteroidales species containing the GA1 ICE, but outcompete their ancestral strain. This event will alter the T6SS sensitive target spectrum and even impact the spatial gut community structure. Besides that, prevalent members of Bacteroidales in human gut were identified to encode acquired interbacterial defense (AID) gene clusters with multiple immunity proteins for defense against T6S-delivered interbacterial competition.39 The AID system acquisition will confer capacity for Bacteroidale to survive in T6SS mediated killing and may help sustain the diversity of community. In a healthy human gut, Enterobacteriales only account for less than 0.1% of microbiota40, but E. coli could be present in the gut microbiome of virtually all adult individuals.41 The first T6SS-dependent antibacterial event of E. coli was identified in pathogenic E. coli strain EAEC 17–2 and APEC TW-XM. Researchers have proven the T6SS dependent antagonistic activity against neighboring nonpathogenic E. coli.42,43 A recent study report that the mouse gut commensal E. coli Mt1B1 employs T6SS to outcompete pathogenic Citrobacter rodentium for niche defense, suggesting a T6SS-dependent competition potential for Enterobacteriales in the gut.44
Collectively, T6SS appears to be important for niche occupancy and sustaining gut microbiome diversity by promoting inter- or intra-species competition. The inherent antagonistic activity can be utilized as a tool to counteract target bacteria in human gut, particularly against certain potential pathogens (Figure 1). For example, introducing T6SS encoding non-toxigenic B. fragilis will eliminate the enterotoxigenic B. fragilis in mouse model.45 Effective T6SS dependent antagonism of pathogenic C. rodentium by gut commensal E. coli may enhance its application value.
The type VII secretion system
The Type VII secretion system (T7SS), also known as the Esx secretion system, is a secretion system specific to Gram-positive bacteria, first identified in pathogenic Mycobacterium tuberculosis.46 M. tuberculosis and M. marinum both utilize the T7SS to deliver virulence factors into the extracellular milieu.47 T7SSs are divided into two categories, Type VIIa system (T7SSa) and Type VIIb system (T7SSb), based on differences in core structural apparatus and exported substrates.48 T7SSa is intensively studied in mycobacteria and is widely distributed in the Actinobacteria phylum, while T7SSb was initially characterized in Staphylococcus aureus and found in various Firmicutes including Bacillus and Listeria species in addition to Staphylococcus.49–52 Mycobacteria can encode up to five homologous but functionally distinct T7SSa systems, designated as ESX-1 to ESX-5, which play various roles in bacterial physiology and virulence.53 Meanwhile, T7SSb is involved in pathogenesis and interbacterial competition by exporting small toxins.54–56
T7SS is functionally similar to the T6SS of Gram-negative bacteria, utilizing effector-immunity (EI) repertoires to exert antagonistic effects through contact-dependent mechanisms (Figure 2). Mougous and colleagues demonstrate that Streptococcus intermedius export LXG toxins through Esx secretion pathway to compete with diverse Firmicute species.57 Interestingly, Firmicutes metagenome screening showed that the LXG protein-encoding genes are widely distributed among many Firmicutes bacterial classes which are mainly adapted to the mammalian gut environment. Bacillus subtilis, a normal gut commensal, encodes six LGX proteins that induce growth inhibition when overexpressed in E. coli.58,59 Moreover, recent research has confirmed that B. subtilis can outcompete its T7SS mutant sister cells in a T7SSb dependent manner.60 Based on these findings, we speculate that Esx system-mediated interbacterial antagonism may contribute to the corresponding Firmicutes species that become dominant in the gut microbial community by eliminating the sensitive target in the same ecological niches. However, in the few Firmicutes species that have been investigated, T7SS seems to be strictly regulated at the transcriptional or posttranslational level and may be activated under specific conditions.61,62 For example, phage predation can induce the transcription of T7SS genes in Enterococcus faecalis, leading to bactericidal activity against gut commensals or pathogens, including E. faecium, S. aureus, and L. monocytogenes.63 A study by Chatterjee and colleagues discovered that phage infection and sub-lethal antibiotic exposure can activate T7SS expression, subsequently triggering T7SS antibacterial activity in E. faecium.64 Exposure to hemin enhances T7SS transcription and effector secretion in S. aureus.65 These studies suggest that T7SS-mediated bacterial competition may play a significant role in preserving community stability under stressful conditions. Additionally, T7SSb is usually restricted to some Firmicutes species inhabiting in gut, suggesting a critical role of T7SS in shaping Firmicutes-rich bacterial communities by competing with other Gram-positive constituents in the gut.57,66
Bacteriocins
Bacteriocins are a promising group of ribosomally synthesized antimicrobial proteins or peptides produced by certain microorganisms.67,68 Many bacterial species have been demonstrated to produce bacteriocins for self-preservation and competition within their ecological niches. These molecules can be produced by both Gram-positive and Gram-negative bacteria, as well as many archaea. Bacteriocins employ distinct mechanisms to attack target bacteria, such as pore formation, inhibition of peptidoglycan synthesis, and interference with gene expression and protein synthesis (Figure 2).68,69 Currently, several hundred bacteriocins have been identified. Some bacteriocins, including many enterobacterial toxins, typically exhibit narrow antimicrobial activity. In contrast, certain other bacteriocins, particularly small peptides derived from Gram-positive bacteria, have a broader spectrum of activity.67,70
The human gut microbiome serves as a rich source of bioactive bacteriocins.71 Bacteriocin production by gut commensals can either encourage or prevent the invasion of new bacterial strains into the community. Additionally, it can play a role in shaping the composition of microbiome members within their ecological niches.67 A majority of human gut microbiome members have been observed to secrete one or more types of antagonistic bacteriocins. Examples include Lactobacillus spp., E. coli, B. subtilis, Enterococcus spp., and Bacteroides spp. As the most predominant bacterial phyla in the human gut, Firmicutes and Bacteroidetes produce the largest number of known bacteriocins, with Actinobacteria and Proteobacteria contributing to a major part of the remaining taxa.72,73
Bacteroidetes encode toxins containing the membrane attack complex/perforin (MACPF) domain. Currently, four Bacteroidales-secreted antimicrobial protein (BSAP) toxins (BSAP-1, BSAP-2, BSAP-3, and BSAP-4) have been identified as mediators of intraspecies competition.74–77 Besides, B. fragilis is able to secrete an eukaryotic-like ubiquitin protein to compete with other B. fragilis strains.78 Bacteroidetocins are another type of peptide toxins produced by diverse members of the Bacteroidetes phylum, which show a broader range of targets, including Bacteroides, Parabacteroides, and Prevotella species.79,80 However, as of now, there is still a lack of in vivo experimental evidence to confirm their antagonistic effects.
Colicins, extensively studied bacteriocins produced by Enterobacteriaceae, are lethal for related bacterial species but not effective against the producing bacteria due to the presence of neutralizing immunity proteins81. These high-molecular-weight toxins attach to specific receptors in the outer membrane of susceptible cells and kill targets through pore formation and nuclease activity.82 Over 26 different types of colicins have been described in E. coli strains to date.81–83–85 Several studies have highlighted the significance of colicin production for the stable colonization of E. coli in the gastrointestinal tract and for survival during intestinal competition.86–88 Colicin-producing strains cannot coexist with the sensitive strains and resistant strains in a liquid culture media, while all phenotypes could coexist in natural gut ecosystem.82,89 That’s possibly due to the spatial isolation between producer strains and sensitive strains. Besides that, colicin synthesis is typically repressed under normal conditions and regulated by signals indicating DNA damage or nutrient limitation.17,90 A study by Margaret A. Riley and colleagues confirmed that the coexistence of colicin-producing and sensitive strains is possible when producers are in a clumped spatial distribution82. Nutrient limitation or colicin induction through lethal toxins enables colicin-producing bacteria to compete and safeguard their niche against invaders. These findings further propose that competitively interacting populations, distributed spatially, can mutually exclude one another, maintaining steady-state coexistence in the ecosystem. This mechanism may foster microbial diversity in the environment.91
Microcins are low-molecular-weight compounds (less than 10 kD) classified as narrow-spectrum bacteriocins. They are mainly derived from E. coli and exhibit receptor-mediated antibacterial activity against Gram-negative bacteria. Although the antimicrobial activity of microcins has been well investigated in vitro, their role in vivo remains unclear92,93. A study by Manuela Raffatellu et al. provided evidence that E. coli Nissle 1917 microcins mediate competition against commensal E. coli in the inflamed gut. This competition could potentially reduce enterobacterial blooms and maintain diversity in the intestinal microbiota.94
Firmicutes, which include Bacillus, Enterococcus, Streptococcus, Staphylococcus, and many other prevalent gut commensals, are capable of producing various posttranslationally modified small peptide bacteriocins known as lantibiotics. Lantibiotics exert their bactericidal activity through pore formation or by preventing cell wall biosynthesis.95 Nisin, the most extensively studied lantibiotic, demonstrates antimicrobial activity against a broad range of Gram-positive bacteria and is currently of interest for clinical applications. Oral administration of nisin can reshape the Firmicutes and Proteobacteria abundance of gut microbiota in animal models.96–98 Lactococcus lactis, which produces nisin F, has been found to stabilize the bacterial population in the gastrointestinal tract of mice.99 Gut-derived Streptococcus salivarius exhibits a narrower spectrum of activity against Fusobacterium nucleatum through the secretion of nisin G.100 Additionally, a novel probiotic bacterium, Lactobacillus plantarum P-8, produces plantaricin, which may contribute to a direct shift in the microbiota structure in the human gut.101 In the gut environment, bacteriocin production will confer a colonization advantage for invading a bacteriocin sensitive ecosystem (Figure 1). Correspondingly, the bacteriocin producer dominant community will prevent sensitive strain colonization. In recent years, many studies have revealed the protective effect of bacteriocins against different intestinal pathogens including Clostridium difficile, Staphylococcus aureus and Salmonella enteritidis etc, Hence, the administration of bacteriocin-producing commensal may become a means for inhibiting potentially problematic bacteria in gut without disrupting the overall structure of the gut microbiota.102 The antibacterial spectrum of bacteriocins secreted by different bacteria varies; some are broad-spectrum, such as nisin, while others are narrow-spectrum, like microcin. Targeted intake of bacteria that secret bacteriocins with specific antibacterial spectrum, holds the potential to selectively adjust the structure of the intestinal microbiota. This modulation may promote conditions conducive to host health.
Exploitation competition
In addition to engaging in direct competition using various tactics such as secretion systems or bacteriocins to harm each other, mammalian gut microbes also partake in indirect competition for shared resources. This type of competition is known as exploitation competition. In exploitative competition, organisms, particularly those with similar niche preferences, can limit their competitors by either consuming finite nutrients more effectively or by releasing molecules that consume nutrients14,103(Figure 2). Advances in metagenomics and metabolomics have enabled scientists to identify species compositions and potential interactions within microbiomes in the same ecosystem. A range of experimental assessments and mathematical modeling have characterized these exploitative interactions among gut microbiome communities.104,105 A metagenomics-based study indicated that pairs of species with the same nutritional profiles exhibit antagonistic behavior as they compete for a similar niche within the human gut.106 For instance, Clostridium ASF356 and Parabacteroides ASF519 compete for glucose, with the former consuming a larger number of compounds in the community than the latter107. Streptococcus oralis and Streptococcus gordonii, sharing the same niche, display intense metabolic competition106. Interestingly, the metabolic competition index, determined by species’ nutritional profiles, typically shows a positive correlation with the co-occurrence of species that compete for the same nutrients, suggesting that the intestinal microbiome is significantly influenced by habitat filtering.106 Bacteroides, a dominant genus in the human microbiota, possesses a broad repertoire of carbohydrate-active enzymes (CAZymes) that help digest polysaccharides from the diet. This enzyme diversity can lead to competition between species.108 For example, various Bacteroides species compete for resources like inulin and xylan in co-culture experiments.109 In the mouse gut, Bacteroides caccae has been observed to out-compete Bacteroides thetaiotaomicron for inulin110. Another example involves the competition between a Firmicutes species, Roseburia intestinalis, and a Bacteroides species, both being efficient xylan degraders. However, R. intestinalis can outcompete B. ovatus during co-culturing in a medium supplemented with xylooligosaccharides.111 Exploitative competition is common among organisms that share overlapping nutrient sources in the gut community, and it clearly affects population and community dynamics (Figure 1). In the gut environment, the competition phenotypes are not fixed. At low population densities, the competition within the community is primarily exploitative, as organisms compete for shared resources.112 However, as the population density increases and resource levels decrease, this interaction shifts toward interference competition. In microbial communities, both exploitative and interference competition can coexist, and the relative importance of each type of competition can vary depending on the specific ecological context and the characteristics of the competing species.
Positive interactions in gut
Besides competing for nutrients, bacteria within the same ecosystem can also engage in positive interactions that mutually benefit each other. Some studies have suggested that competition dominates microbial species interactions, and positive interactions are much less common than competition in microbial ecosystems.113,114 However, metabolic dissimilarity between co-occurring microorganisms could provide a survival advantage through complementary biosynthetic capabilities, suggesting a cooperation feature between microbes115. In recent years, both empirical and theoretical research has increasingly indicated that positive interactions among microbes are prevalent and could play significant roles in growth, composition, and the shaping of microbial communities.16–116–117,118 Positive interactions are defined as instances where at least one partner involved in the interaction experiences an improvement in fitness. Since interactions are often studied between pairs of microorganisms, we will also present them in pairs in the following sections. These interactions enhance the fitness of partners either unidirectionally or bidirectionally by generating beneficial metabolites, such as amino acids119,120, vitamins121 or products from polymer degradation122. Generally, positive interactions between gut microbes fall into three main types: commensalism, mutualism, and cooperation105.
Commensalism
Commensalism is a unidirectional positive interaction between two species, wherein one species experience increased benefits without causing any harm or providing benefit to the other species (Figure 3). Culture-based studies suggested that commensalisms were the most common positive interactions in bacterial community123. Dietary polysaccharides serve as a primary nutrient source for gut microbes, directly influencing the composition and metabolism of the microbiota124. The Bacteroidetes phylum is renowned for its ability to degrade a wide range of polysaccharides into monosaccharides, oligosaccharides, or other fermentable end products, which can be utilized by other members of the microbial community.125 This complex glycans utilization ability is typically facilitated by polysaccharide utilization loci (PULs), which encompass glycan-binding proteins, oligosaccharide transporter proteins, carbohydrate sensors, and CAZymes.126 The strong polysaccharide utilization ability of Bacteroides enriches its own abundance and also provide various nutrients for other bacteria in the community (Figure 1). For example, Bifidobacterium animalis utilizes xylooligosaccharides, which are xylan hydrolysis products from Bacteroides, as a carbon source127. In a rat gut model, inulin fermentation by B. uniformis produces fructo-oligosaccharides and monosaccharides, which further support the growth of Blautia glucerasea, Clostridium indolis, and Bifidobacterium animalis.128
Figure 3.
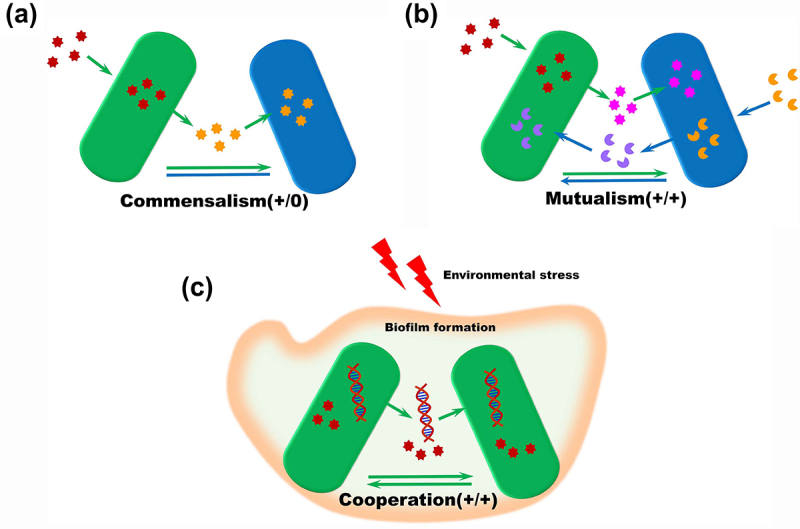
Types of positive interactions within gut microbiome. positive interactions are common and important in a population. One species can utilize metabolites produced by another species (a), while different bacteria species can also cross-feed each other by exchanging metabolites(b). (c) bacteria sister cells could provide cross-protection by HGT, biofilm formation and cross feeding response to environmental stressors such as antibiotics or nutrient deficiencies.
Among the first microbes to colonize the human gastrointestinal tract, Bifidobacteria are the most abundant bacteria in the infant gut and are believed to confer positive health benefits to their host. Bifidobacterium species also possess a diverse array of CAZymes that enable them to break down diet-derived glycans (e.g., starch and xylan) as well as host-derived carbohydrates like mucin and HMOs (Human Milk Oligosaccharides).129,130 The resulting fermented products, such as sialic acid and glucose, are subsequently utilized by other members of the Bifidobacterium genus.131 In vitro studies have also demonstrated that the 1,2-propanediol produced by Bifidobacterium breve can enhance the growth of Lactobacillus reuteri, and metabolites like acetate and lactate from Bifidobacterium adolescentis can be used by Eubacterium hallii.132,133 The gut microbes, which has limited capacities for processing dietary and host-derived polysaccharides, typically relies on the fermentation products of other organisms capable of fermenting polysaccharides. Therefore, dietary glycans could increase the abundance of glycan-utilizing organisms and also influence the overall diversity and structure of the gut microbiome through positive microbe interactions.
Mutualism
Mutualism involves a bidirectional positive interaction between two distinct species, and it is widely observed in natural ecological communities134,135 (Figure 3). In recent years, an increasing number of studies have been focusing on positive interactions among microbes and have defined similar interactions using various terms, such as syntropy, mutual/bidirectional cross-feeding, mutualism, and synergism, among others.136–139 Mutualism refers to the exchange of metabolic products between individuals of different species for the benefit of both.138
Within the complex ecological community of the gut microbiota, numerous positive mutual interactions can be found. For example, Bifidobacterium longum and Eubacterium rectale are prevalent species in the human colon microbiota. When co-cultured on Arabinoxylan oligosaccharides (AXOS), B. longum NCC2705 consumes the arabinose substituents of AXOS and produces acetate, which serves as a substrate for E. rectale ATCC 33,656 to generate butyrate. Simultaneously, E. rectale releases xylose through extracellular AXOS degradation, further supporting the production of acetate by B. longum.137 A study by Thi Phuong Nam Bui et al. reported that butyrate-producing bacterium Anaerostipes rhamnosivorans produces H2/CO2, which can be utilized by acetogenic B. hydrogenotrophica to form acetate. This acetate, in turn, is used by A. rhamnosivorans to initiate the conversion of lactate to butyrate.140 In a gnotobiotic mouse model, the archaeal representative Methanobrevibacter smithii enhances B. thetaiotaomicron fermentation of fructans, resulting in acetate production. In reciprocation, M. smithii benefits from formate produced by B. thetaiotaomicron, which supports methanogenesis.141 Beyond the mutual exchange of metabolites, mutualism can also involve the collaboration of different species to form a biofilm138,142. Gut commensals, including B. bifidum, B. longum subsp. infantis, P. distasonis, and B. ovatus, can create substantial biofilms when co-cultured in various combinations143, suggesting cooperative interactions in the formation of multispecies biofilms. However, the underlying molecular mechanisms and the specific roles of each strain in these cooperative effects still need to be determined. Mutual cross-feeding interactions can expand the metabolic niches of the interacting individuals, promoting microbial diversity and preventing competitive exclusion.144,145 Mutualistic trade-offs of essential metabolites, like amino acids, can reduce the biosynthetic burden for the utilization of metabolic pathways.146
Cooperation
As discussed earlier, mutualism and commensalism involve interactions between distinct species, whereas cooperation described here implies an intraspecies interaction that enhances the fitness of neighboring cells sharing a specific genotype (Figure 3). Microbial cooperation within a single organism can bolster microbial tolerance to environmental and ecological stressors, such as antibiotics. A study by Yurtsev et al. demonstrated that co-culturing two resistant E. coli strains can lead to effective cross-protection, where they shield each other in a multidrug environment that inhibits the growth of either strain alone.147 Similarly, a pair of amino acid auxotrophic E. coli strains can complement each other’s deficiencies in a co-culture, demonstrating a cross-feeding cooperation effect.148
Within the human gut microbiome, horizontal gene transfer (HGT) events involving antibiotic-resistance genes (ARGs) are frequently observed among commensals and opportunistic pathogens (e.g., E. coli and K. pneumoniae, E. faecalis and E. faecium).149 HGT of beneficial genes in the gut microbiome is also considered a form of cooperation.150 It is common for microbes to gain or lost genes. Inter-bacterial HGT enables the acquisition of potentially adaptive genes to the recipient cells to help them survive in stressful environments (Figure 3). The formation of biofilms by prokaryotic organisms often necessitates significant cooperation to share extracellular polymeric substances among cells. Cells attached to a biofilm surface exhibit different phenotypes compared to their planktonic state, displaying stronger colonization abilities and greater tolerance to external stressors. Convincing evidence suggests that many gut bacterial species form biofilms on mucosal surfaces to enhance the efficiency of complex polysaccharide degradation and increase tolerance to environmental stressors.151,152 Gut molecules like bile have been shown to induce biofilm formation in gut bacteria, including Bacteroides, Lactobacillus, and Bifidobacterium, underscoring the potential importance of biofilm formation as a colonization-related factor for gut bacteria.153 Cooperation events are more prominent when the microorganisms are confronted with various environmental pressures, including antibiotics and nutrient deficiencies. Such interactions may carry substantial ecological importance in upholding gut species stability when exposed to external stresses.
Understanding of the positive interaction networks based on nutrients utilization is crucial for developing a microbiota targeting diet to increase the abundance of beneficial gut microbes. Providing specific dietary fibers tailored toward enrichment of particular gut microbial member corresponding to their enzymatic profile could be an option to obtain predictable change in microbial composition. For example, administration of acetylated galactoglucomannan (AcGGM) fiber specifically aligned with enzymatic capabilities of butyrate-producing species Roseburia and Faecalibacterium increased the relative abundance of Faecalibacterium and specific phylotypes of Roseburia in porcine gut.154 Meanwhile, abundance of a group of non-fiber-degrading taxa are also elevated, largely dependent on cross-feeding events. To leverage positive interactions among microbial communities and precisely regulate the gut microbiota, such as through exogenous dietary fibers, for optimizing host health, a deeper understanding of the complex interplay between gut microbial communities is required.
Membrane vesicles: an interaction mechanism exhibiting dual functions
Within the gut microbial ecosystem, the balance between competition and cooperation among microorganisms is dynamic and greatly influenced by environmental factors. To navigate through complex and ever-changing environments, bacteria have evolved a dual functional mechanism, MVs, to interact bidirectionally with their community. This device enables the delivery of a wide array of cargoes, including nucleic acids, quorum sensing signals, toxins, and valuable nutrients, facilitating both competitive interactions and the acquisition of beneficial effects. This adaptability allows bacteria to thrive in intricate environments.
MVs are bubble-like structures originating from the outer membranes of Gram-negative bacteria or cytoplasmic membrane vesicles formed by endolysin-triggered cell lysis in Gram-positive bacteria155 (Figure 2). Proteomic and biochemical analyses have revealed that these small particles typically transport microbial substances, including nucleic acids, proteins, lipids, and metabolites, that impact diverse biological processes such as quorum sensing (QS), biofilm formation, and cell-to-cell communication156. Over the past decade, the majority of studies have focused on cytotoxic factors delivered by MVs in Gram-negative pathogens or commensals to manipulate the host immune response157. However, in this context, we will delve into the activity of gut bacterial MVs against different bacterial species within gut commensals.
Gut microbes could secrete active compounds contained within MVs into the intestinal lumen, influencing other commensals that are at a distance from their parent cells. Due to their cargo diversity, MVs exhibit various functions, from bacterial competition to nutrient utilization and even stress resistance.158 Recent studies have indicated that the potential mechanism for delivering bacteriocins to target cells is through MVs. For instance, B. fragilis antimicrobial peptide BSAP-1 secretion is mediated by MVs.74 Proteomic analyses of Lactobacillus-derived MVs showed that MVs could serve as vehicles for delivering antimicrobial peptides,159 a finding further confirmed in a strain of Lactobacillus acidophilus by another study.160 MVs originating from gut Myxococcus xanthus have demonstrated the ability to kill E. coli cells.161 However, more evidence is needed to firmly establish the competitive role of microbiota-derived MVs in the gut.
Beyond facilitating competitive interactions among bacterial community members, gut microbiota MVs also promote positive interactions by releasing enzymes into the intestinal lumen. Proteomic analysis of outer membrane vesicles (OMVs) in B. fragilis and B. thetaiotaomicron has identified sugar hydrolases and proteases that are preferentially packaged within the MVs.162 Specific Bacteroides strains also deliver sulfatases through MVs to aid in the degradation of mucin glycans, thereby providing nutrients to other community members.163 Hence, the enzymes enclosed within OMVs act as “public good” that provide benefits to the entire bacterial community. Additionally, besides delivering crucial enzymes for glycan degradation, MVs release enzymes that confer antibiotic resistance. MVs produced by B. thetaiotaomicron and several other species, which carry β-lactamases, can shield susceptible commensal bacteria from β-lactam antibiotics within the microbial community164. Furthermore, membrane vesicle-mediated HGT can facilitate the transfer of functional ARGs between bacterial species.165
Conclusion and future perspective
Bacterial interactions play a pivotal role in shaping the diversity and stability of the gut microbiota. Nevertheless, this intricate and dynamic system is continually influenced by a broad range of factors, including diet, medication (especially antibiotics), infections, stress, and the overall health status of the host. Despite decades of research on microbial interactions, our comprehension of these interactions within natural communities remains limited. Through comprehensive and systematic investigations into the interplay of gut bacteria, both in vivo and in vitro, we can unveil the complex mechanisms that underlie gut microbiota stability and diversity. It is evident that we still have much to uncover about microbial interactions. The ongoing exploration of this area is imperative for advancing our understanding of the gut microbiota and for developing novel strategies to modulate it for therapeutic purposes.
Acknowledgments
This work was supported by the National Natural Science Foundation of China fund (NSFC 31700126) and Heilongjiang Postdoctoral Scientific Research Developmental Fund (LHB-Q21153) to H.B., a National Natural Science Foundation of China fund (NSFC82020108022) to SL.L., and a Heilongjiang Provincial Natural Science Foundation of China (LH2023H005) and a Heilongjiang Postdoctoral Scientific Research Developmental Fund to S.W. (LBH-Q20149).
Funding Statement
The work was supported by the Heilongjiang Provincial Natural Science Foundation of China [LH2023H005]; Heilongjiang Postdoctoral Scientific Research Developmental Fund [LBH-Q20149]; Heilongjiang Postdoctoral Scientific Research Developmental Fund [LHB-Q21153]; National Natural Science Foundation of China fund [NSFC31700126]; National Natural Science Foundation of China fund [NSFC82020108022].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Lynch SV, Pedersen O, Phimister EG.. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–18. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Sci. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Marchesi JR. Prokaryotic and eukaryotic diversity of the human gut. Adv Appl Microbiol. 2010;72:43–62. [DOI] [PubMed] [Google Scholar]
- 4.Dominguez-Bello MG, Godoy-Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. 2019;68(6):1108–1114. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Sci. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, Segata N, Bork P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28(4):561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machado D, Maistrenko OM, Andrejev S, Kim Y, Bork P, Patil KR, Patil KR. Polarization of microbial communities between competitive and cooperative metabolism. Nat Ecol Evol. 2021;5(2):195–203. doi: 10.1038/s41559-020-01353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith HR, Srimani JK, Lee AJ, Lopatkin AJ, You L. Collective antibiotic tolerance: mechanisms, dynamics and intervention. Nat Chem Biol. 2015;11(3):182–188. doi: 10.1038/nchembio.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer MA, Kainz K, Carmona-Gutierrez D, Madeo F. Microbial wars: competition in ecological niches and within the microbiome. Microb Cell. 2018;5(5):215–219. doi: 10.15698/mic2018.05.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghoul M, Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24(10):833–845. doi: 10.1016/j.tim.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2(1):589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 16.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533(7602):255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11(4):285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 18.Green ER, Mecsas J, Kudva IT. Bacterial secretion systems: an overview. Microbiol Spectr. 2016;4(1). doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman AS, Mauer TJ, Forest KT, Goodrich-Blair H. A widespread bacterial secretion system with diverse substrates. mBio. 2021;12(4):e0195621. doi: 10.1128/mBio.01956-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorasia DG, Veith PD, Reynolds EC. The type IX secretion system: advances in structure, function and organisation. Microorgan. 2020;8(8):1173. doi: 10.3390/microorganisms8081173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer T, Finney AJ, Saha CK, Atkinson GC, Sargent F. A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol Microbiol. 2021;115(3):345–355. doi: 10.1111/mmi.14599. [DOI] [PubMed] [Google Scholar]
- 22.Bao H, Wang S, Zhao JH, Liu SL. Salmonella secretion systems: differential roles in pathogen-host interactions. Microbiol Res. 2020;241:126591. doi: 10.1016/j.micres.2020.126591. [DOI] [PubMed] [Google Scholar]
- 23.Crisan CV, Hammer BK. The vibrio cholerae type VI secretion system: toxins, regulators and consequences. Environ Microbiol. 2020;22(10):4112–4122. doi: 10.1111/1462-2920.14976. [DOI] [PubMed] [Google Scholar]
- 24.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. Secretion systems in gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13(6):343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 25.Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol. 2009;99(Suppl S1):S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulthurst S. The type VI secretion system: a versatile bacterial weapon. Microbiol. 2019;165(5):503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 27.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475(7356):343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113(13):3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. Bmc Genom. 2016;17(1):58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE, Mekalanos J. Evidence of extensive DNA transfer between bacteroidales species within the human gut. mBio. 2014;5(3):e01305–01314. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coyne MJ, Comstock LE, Sandkvist M, Cascales E, Christie PJ. Type VI secretion systems and the gut microbiota. Microbiol Spectr. 2019;7(2). doi: 10.1128/microbiolspec.PSIB-0009-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host & Microbe. 2017;22(3):411–419 e414. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A. 2016;113(13):3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robitaille S, Simmons EL, Verster AJ, McClure EA, Royce DB, Trus E, Swartz K, Schultz D, Nadell CD, Ross BD. Community composition and the environment modulate the population dynamics of type VI secretion in human gut bacteria. Nat Ecol Evol. 2023;7(12):2092–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Bayona L, Coyne MJ, Comstock LE, Blokesch M. Mobile type VI secretion system loci of the gut bacteroidales display extensive intra-ecosystem transfer, multi-species spread and geographical clustering. PLoS Genet. 2021;17(4):e1009541. doi: 10.1371/journal.pgen.1009541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao H, Coyne MJ, Garcia-Bayona L, Comstock LE, O’Toole G. Analysis of effector and immunity proteins of the GA2 type VI secretion systems of gut bacteroidales. J Bacteriol. 2022;204(7):e0012222. doi: 10.1128/jb.00122-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheahan ML, Coyne MJ, Flores K, Garcia-Bayona L, Chatzidaki-Livanis M, Sundararajan A, Holst AQ, Barquera B, Comstock LE. A ubiquitous mobile genetic element disarms a bacterial antagonist of the gut microbiota. bioRxiv. 2023. [Google Scholar]
- 39.Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar AM, Peterson SB, Borenstein E, Mougous JD. Human gut bacteria contain acquired interbacterial defence systems. Nature. 2019;575(7781):224–228. doi: 10.1038/s41586-019-1708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahrodia T, Das S, Bakshi S, Das B. Structure, functions, and diversity of the healthy human microbiome. Prog Mol Biol Transl Sci. 2022;191:53–82. [DOI] [PubMed] [Google Scholar]
- 41.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8(3):207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 42.Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H, McCormick BA. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun. 2014;82(9):3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013;3(1):36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Serapio-Palacios A, Woodward SE, Vogt SL, Deng W, Creus-Cuadros A, Huus KE, Cirstea M, Gerrie M, Barcik W, Yu H, et al. Type VI secretion systems of pathogenic and commensal bacteria mediate niche occupancy in the gut. Cell Rep. 2022;39(4):110731. doi: 10.1016/j.celrep.2022.110731. [DOI] [PubMed] [Google Scholar]
- 45.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17(9):1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178(5):1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5(11):883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 48.Bowman L, Palmer T. The type VII secretion system of staphylococcus. Annu Rev Microbiol. 2021;75(1):471–494. doi: 10.1146/annurev-micro-012721-123600. [DOI] [PubMed] [Google Scholar]
- 49.Pallen MJ. The ESAT-6/WXG100 superfamily – and a new gram-positive secretion system? Trends Microbiol. 2002;10(5):209–212. doi: 10.1016/S0966-842X(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 50.Bottai D, Groschel MI, Brosch R. Type VII secretion systems in gram-positive bacteria. Curr Top Microbiol Immunol. 2017;404:235–265. [DOI] [PubMed] [Google Scholar]
- 51.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. The enigmatic Esx proteins: looking beyond mycobacteria. Trends Microbiol. 2017;25(3):192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Bowran K, Palmer T. Extreme genetic diversity in the type VII secretion system of listeria monocytogenes suggests a role in bacterial antagonism. Microbiol. 2021;167(3). doi: 10.1099/mic.0.001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bunduc CM, Bitter W, Houben ENG. Structure and function of the mycobacterial type VII secretion systems. Annu Rev Microbiol. 2020;74(1):315–335. doi: 10.1146/annurev-micro-012420-081657. [DOI] [PubMed] [Google Scholar]
- 54.Ohr RJ, Anderson M, Shi M, Schneewind O, Missiakas D, Silhavy TJ. EssD, a nuclease effector of the staphylococcus aureus ESS pathway. J Bacteriol. 2017;199(1). doi: 10.1128/JB.00528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. The type VII secretion system of staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nature Microbiol. 2016;2(1):16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulhuq FR, Gomes MC, Duggan GM, Guo M, Mendonca C, Buchanan G, Chalmers JD, Cao Z, Kneuper H, Murdoch S, et al. A membrane-depolarizing toxin substrate of the staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc Natl Acad Sci USA. 2020;117(34):20836–20847. doi: 10.1073/pnas.2006110117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife. 2017;6:6. doi: 10.7554/eLife.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holberger LE, Garza-Sanchez F, Lamoureux J, Low DA, Hayes CS. A novel family of toxin/antitoxin proteins in bacillus species. FEBS Lett. 2012;586(2):132–136. doi: 10.1016/j.febslet.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaundal S, Deep A, Kaur G, Thakur KG. Molecular and biochemical characterization of YeeF/YezG, a polymorphic toxin-immunity protein pair from Bacillus subtilis. Front Microbiol. 2020;11:95. doi: 10.3389/fmicb.2020.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tassinari M, Doan T, Bellinzoni M, Chabalier M, Ben-Assaya M, Martinez M, Gaday Q, Alzari PM, Cascales E, Fronzes R, et al. The antibacterial type VII secretion system of bacillus subtilis: structure and interactions of the pseudokinase YukC/EssB. mBio. 2022;13(5):e0013422. doi: 10.1128/mbio.00134-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baptista C, Barreto HC, Sao-Jose C, Cascales E. High levels of DegU-P activate an esat-6-like secretion system in bacillus subtilis. PLoS ONE. 2013;8(7):e67840. doi: 10.1371/journal.pone.0067840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulthess B, Bloes DA, Berger-Bachi B. Opposing roles of σB and σB-controlled SpoVG in the global regulation of esxA in staphylococcus aureus. BMC Microbiol. 2012;12(1):17. doi: 10.1186/1471-2180-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee A, Willett JLE, Nguyen UT, Monogue B, Palmer KL, Dunny GM, Duerkop BA, Hatfull GF. Parallel genomics uncover novel enterococcal-bacteriophage interactions. mBio. 2020;11(2). doi: 10.1128/mBio.03120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatterjee A, Willett JLE, Dunny GM, Duerkop BA, Blokesch M. Phage infection and sub-lethal antibiotic exposure mediate enterococcus faecalis type VII secretion system dependent inhibition of bystander bacteria. PLoS Genet. 2021;17(1):e1009204. doi: 10.1371/journal.pgen.1009204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casabona MG, Kneuper H, Alferes de Lima D, Harkins CP, Zoltner M, Hjerde E, Holden MTG, Palmer T. Haem-iron plays a key role in the regulation of the Ess/type VII secretion system of staphylococcus aureus RN6390. Microbiol (Reading). 2017;163(12):1839–1850. doi: 10.1099/mic.0.000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boopathi S, Liu D, Jia AQ. Molecular trafficking between bacteria determines the shape of gut microbial community. Gut Microbes. 2021;13(1):1959841. doi: 10.1080/19490976.2021.1959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heilbronner S, Krismer B, Brotz-Oesterhelt H, Peschel A. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol. 2021;19(11):726–739. doi: 10.1038/s41579-021-00569-w. [DOI] [PubMed] [Google Scholar]
- 68.Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11(2):95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 69.Garcia-Bayona L, Comstock LE. Bacterial antagonism in host-associated microbial communities. Science. 2018;361:6408. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]
- 70.Sanchez-Hidalgo M, Montalban-Lopez M, Cebrian R, Valdivia E, Martinez-Bueno M, Maqueda M. AS-48 bacteriocin: close to perfection. Cell Mol Life Sci. 2011;68(17):2845–2857. doi: 10.1007/s00018-011-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng J, Ganzle MG, Lin XB, Ruan L, Sun M. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol. 2015;17(6):2133–2143. doi: 10.1111/1462-2920.12662. [DOI] [PubMed] [Google Scholar]
- 72.Lopetuso LR, Giorgio ME, Saviano A, Scaldaferri F, Gasbarrini A, Cammarota G. Bacteriocins and bacteriophages: therapeutic weapons for gastrointestinal diseases? Int J Mol Sci. 2019;20(1):183. doi: 10.3390/ijms20010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drissi F, Buffet S, Raoult D, Merhej V. Common occurrence of antibacterial agents in human intestinal microbiota. Front Microbiol. 2015;6:441. doi: 10.3389/fmicb.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. An antimicrobial protein of the gut symbiont bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol. 2014;94(6):1361–1374. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE, Huffnagle GB. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio. 2016;7(4). doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McEneany VL, Coyne MJ, Chatzidaki-Livanis M, Comstock LE. Acquisition of MACPF domain-encoding genes is the main contributor to LPS glycan diversity in gut bacteroides species. ISME J. 2018;12(12):2919–2928. doi: 10.1038/s41396-018-0244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shumaker AM, Laclare McEneany V, Coyne MJ, Silver PA, Comstock LE, DiRita VJ. Identification of a fifth antibacterial toxin produced by a single bacteroides fragilis strain. J Bacteriol. 2019;201(8). doi: 10.1128/JB.00577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chatzidaki-Livanis M, Coyne MJ, Roelofs KG, Gentyala RR, Caldwell JM, Comstock LE, Mekalanos JJ. Gut symbiont bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio. 2017;8(6). doi: 10.1128/mBio.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matano LM, Coyne MJ, Garcia-Bayona L, Comstock LE, Stephen Trent M. Bacteroidetocins target the essential outer membrane protein BamA of bacteroidales symbionts and pathogens. mBio. 2021;12(5):e0228521. doi: 10.1128/mBio.02285-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coyne MJ, Bechon N, Matano LM, McEneany VL, Chatzidaki-Livanis M, Comstock LE. A family of anti-bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat Commun. 2019;10(1):3460. doi: 10.1038/s41467-019-11494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71(1):158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Majeed H, Gillor O, Kerr B, Riley MA. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 2011;5(1):71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smajs D, Weinstock GM. Genetic organization of plasmid ColJs, encoding colicin js activity, immunity, and release genes. J Bacteriol. 2001;183(13):3949–3957. doi: 10.1128/JB.183.13.3949-3957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Micenkova L, Bosak J, Kucera J, Hrala M, Dolejsova T, Sedo O, Linke D, Fiser R, Smajs D. Colicin Z, a structurally and functionally novel colicin type that selectively kills enteroinvasive Escherichia coli and shigella strains. Sci Rep. 2019;9(1):11127. doi: 10.1038/s41598-019-47488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rendueles O, Beloin C, Latour-Lambert P, Ghigo JM. A new biofilm-associated colicin with increased efficiency against biofilm bacteria. ISME J. 2014;8(6):1275–1288. doi: 10.1038/ismej.2013.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gillor O, Giladi I, Riley MA. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 2009;9(1):165. doi: 10.1186/1471-2180-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, Gerlach RG, Stecher B, Galán JE. Inflammation fuels colicin Ib-dependent competition of salmonella serovar typhimurium and E. coli in enterobacterial blooms. PLoS Pathog. 2014;10(1):e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hancock V, Dahl M, Klemm P. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J Med Microbiol. 2010;59(Pt 4):392–399. doi: 10.1099/jmm.0.008672-0. [DOI] [PubMed] [Google Scholar]
- 89.Gordon DM, Riley MA, Pinou T. Pinou T: temporal changes in the frequency of colicinogeny in Escherichia coli from house mice. Microbiol. 1998;144(8):2233–2240. doi: 10.1099/00221287-144-8-2233. [DOI] [PubMed] [Google Scholar]
- 90.Gillor O, Vriezen JAC, Riley MA. The role of SOS boxes in enteric bacteriocin regulation. Microbiol. 2008;154(Pt 6):1783–1792. doi: 10.1099/mic.0.2007/016139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428(6981):412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- 92.Rebuffat S. Microcins in action: amazing defence strategies of enterobacteria. Biochem Soc Trans. 2012;40(6):1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 93.Yang SC, Lin CH, Sung CT, Fang JY. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol. 2014;5:241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540(7632):280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61(1):477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 96.Jozefiak D, Kieronczyk B, Juskiewicz J, Zdunczyk Z, Rawski M, Dlugosz J, Sip A, Hojberg O, Loh G. Dietary nisin modulates the gastrointestinal microbial ecology and enhances growth performance of the broiler chickens. PLoS ONE. 2013;8(12):e85347. doi: 10.1371/journal.pone.0085347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jia Z, Chen A, Bao F, He M, Gao S, Xu J, Zhang X, Niu P, Wang C. Effect of nisin on microbiome-brain-gut axis neurochemicals by Escherichia coli-induced diarrhea in mice. Microb Pathog. 2018;119:65–71. doi: 10.1016/j.micpath.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 98.O’Reilly C, Grimaud GM, Coakley M, O’Connor PM, Mathur H, Peterson VL, O’Donovan CM, Lawlor PG, Cotter PD, Stanton C, et al. Modulation of the gut microbiome with nisin. Sci Rep. 2023;13(1):7899. doi: 10.1038/s41598-023-34586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Staden DA, Brand AM, Endo A, Dicks LM. Intraperitoneally injected, may have a stabilizing effect on the bacterial population in the gastro-intestinal tract, as determined in a preliminary study with mice as model. Lett Appl Microbiol. 2011;53(2):198–201. doi: 10.1111/j.1472-765X.2011.03091.x. [DOI] [PubMed] [Google Scholar]
- 100.Lawrence GW, McCarthy N, Walsh CJ, Kunyoshi TM, Lawton EM, O’Connor PM, Begley M, Cotter PD, Guinane CM. Effect of a bacteriocin-producing Streptococcus salivarius on the pathogen Fusobacterium nucleatum in a model of the human distal colon. Gut Microbes. 2022;14(1):2100203. doi: 10.1080/19490976.2022.2100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwok LY, Guo Z, Zhang J, Wang L, Qiao J, Hou Q, Zheng Y, Zhang H. The impact of oral consumption of lactobacillus plantarum P-8 on faecal bacteria revealed by pyrosequencing. Benef Microbes. 2015;6(4):405–413. doi: 10.3920/BM2014.0063. [DOI] [PubMed] [Google Scholar]
- 102.Umu OC, Bauerl C, Oostindjer M, Pope PB, Hernandez PE, Perez-Martinez G, Diep DB. The potential of class II bacteriocins to modify gut microbiota to improve host health. PLoS ONE. 2016;11:e0164036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stubbendieck RM, Straight PD, Margolin W. Multifaceted interfaces of bacterial competition. J Bacteriol. 2016;198(16):2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14(6):e8157. doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29(11):R538–R544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci USA. 2013;110(31):12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Biggs MB, Medlock GL, Moutinho TJ, Lees HJ, Swann JR, Kolling GL, Papin JA. Systems-level metabolism of the altered schaedler flora, a complete gut microbiota. ISME J. 2017;11(2):426–438. doi: 10.1038/ismej.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5(7):e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24(1):40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leth ML, Ejby M, Workman C, Ewald DA, Pedersen SS, Sternberg C, Bahl MI, Licht TR, Aachmann FL, Westereng B, et al. Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nature Microbiol. 2018;3(5):570–580. doi: 10.1038/s41564-018-0132-8. [DOI] [PubMed] [Google Scholar]
- 112.Holdridge EM, Cuellar-Gempeler C, terHorst CP. A shift from exploitation to interference competition with increasing density affects population and community dynamics. Ecol Evol. 2016;6(15):5333–5341. doi: 10.1002/ece3.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Foster KR, Bell T. Bell T: competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 114.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112(20):6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.D’Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35(5):455–488. doi: 10.1039/C8NP00009C. [DOI] [PubMed] [Google Scholar]
- 117.Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62(1):375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 118.Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36(5):990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 119.Ziesack M, Gibson T, Oliver JKW, Shumaker AM, Hsu BB, Riglar DT, Giessen TW, DiBenedetto NV, Bry L, Way JC, et al. Engineered interspecies amino acid cross-feeding increases population evenness in a synthetic bacterial consortium. mSystems. 2019;4(4). doi: 10.1128/mSystems.00352-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ponomarova O, Gabrielli N, Sevin DC, Mulleder M, Zirngibl K, Bulyha K, Andrejev S, Kafkia E, Typas A, Sauer U, et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017;5(4):345–357.e6. doi: 10.1016/j.cels.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sokolovskaya OM, Shelton AN, Taga ME. Sharing vitamins: cobamides unveil microbial interactions. Sci. 2020;369(6499):6499. doi: 10.1126/science.aba0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Solden LM, Naas AE, Roux S, Daly RA, Collins WB, Nicora CD, Purvine SO, Hoyt DW, Schuckel J, Jorgensen B, et al. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nature Microbiol. 2018;3(11):1274–1284. doi: 10.1038/s41564-018-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kehe J, Ortiz A, Kulesa A, Gore J, Blainey PC, Friedman J. Positive interactions are common among culturable bacteria. Sci Adv. 2021;7(45):eabi7159. doi: 10.1126/sciadv.abi7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alvarez-Mercado AI, Plaza-Diaz J. Dietary polysaccharides as modulators of the gut microbiota ecosystem: an update on their impact on health. Nutrients. 2022;14(19):4116. doi: 10.3390/nu14194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McKee LS, La Rosa SL, Westereng B, Eijsink VG, Pope PB, Larsbrink J. Polysaccharide degradation by the bacteroidetes: mechanisms and nomenclature. Environ Microbiol Rep. 2021;13(5):559–581. doi: 10.1111/1758-2229.12980. [DOI] [PubMed] [Google Scholar]
- 126.Grondin JM, Tamura K, Dejean G, Abbott DW, Brumer H, O’Toole G. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199(15). doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeybek N, Rastall RA, Buyukkileci AO. Utilization of xylan-type polysaccharides in co-culture fermentations of bifidobacterium and bacteroides species. Carbohydr Polym. 2020;236:116076. doi: 10.1016/j.carbpol.2020.116076. [DOI] [PubMed] [Google Scholar]
- 128.Tannock GW, Lawley B, Munro K, Sims IM, Lee J, Butts CA, Roy N, Macfarlane GT. RNA–stable-Isotope probing shows utilization of carbon from inulin by specific bacterial populations in the rat large bowel. Appl Environ Microbiol. 2014;80(7):2240–2247. doi: 10.1128/AEM.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5(1):15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018;26(4):339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 131.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D, Macfarlane GT. Metabolism of sialic acid by bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80(14):4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cheng CC, Duar RM, Lin X, Perez-Munoz ME, Tollenaar S, Oh JH, van Pijkeren JP, Li F, van Sinderen D, Ganzle MG, et al. Ecological importance of cross-feeding of the intermediate metabolite 1,2-propanediol between bacterial gut symbionts. Appl Environ Microbiol. 2020;86(11). doi: 10.1128/AEM.00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. Two routes of metabolic cross-feeding between bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72(5):3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thebault E, Fontaine C. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science. 2010;329(5993):853–856. doi: 10.1126/science.1188321. [DOI] [PubMed] [Google Scholar]
- 135.Rohr RP, Saavedra S, Bascompte J. Ecological networks. On the structural stability of mutualistic systems. Science. 2014;345(6195):1253497. doi: 10.1126/science.1253497. [DOI] [PubMed] [Google Scholar]
- 136.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Riviere A, Gagnon M, Weckx S, Roy D, De Vuyst L, Schloss PD. Mutual cross-feeding interactions between bifidobacterium longum subsp. longum NCC2705 and eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol. 2015;81(22):7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10(8):538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 139.Schink B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek. 2002;81(1–4):257–261. doi: 10.1023/A:1020579004534. [DOI] [PubMed] [Google Scholar]
- 140.Bui TPN, Schols HA, Jonathan M, Stams AJM, de Vos WM, Plugge CM. Mutual metabolic interactions in co-cultures of the intestinal anaerostipes rhamnosivorans with an acetogen, methanogen, or pectin-degrader affecting butyrate production. Front Microbiol. 2019;10:2449. doi: 10.3389/fmicb.2019.02449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103(26):10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Palmer RJ Jr., Kazmerzak K, Hansen MC, Kolenbrander PE, Moore RN. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69(9):5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sadiq FA, Wenwei L, Heyndrickx M, Flint S, Wei C, Jianxin Z, Zhang H. Synergistic interactions prevail in multispecies biofilms formed by the human gut microbiota on mucin. FEMS Microbiol Ecol. 2021;97(8). doi: 10.1093/femsec/fiab096. [DOI] [PubMed] [Google Scholar]
- 144.Ona L, Giri S, Avermann N, Kreienbaum M, Thormann KM, Kost C. Obligate cross-feeding expands the metabolic niche of bacteria. Nat Ecol Evol. 2021;5(9):1224–1232. doi: 10.1038/s41559-021-01505-0. [DOI] [PubMed] [Google Scholar]
- 145.Hoek M, Merks RMH. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. BMC Syst Biol. 2017;11(1):56. doi: 10.1186/s12918-017-0430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111(20):E2149–2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yurtsev EA, Conwill A, Gore J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc Natl Acad Sci USA. 2016;113(22):6236–6241. doi: 10.1073/pnas.1523317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2014;8(5):953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McInnes RS, McCallum GE, Lamberte LE, van Schaik W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr Opin Microbiol. 2020;53:35–43. doi: 10.1016/j.mib.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 150.Lee IPA, Eldakar OT, Gogarten JP, Andam CP. Bacterial cooperation through horizontal gene transfer. Trends Ecol Evol. 2022;37(3):223–232. doi: 10.1016/j.tree.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 151.Macfarlane S, Macfarlane GT. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl Environ Microbiol. 2006;72(9):6204–6211. doi: 10.1128/AEM.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dubois T, Tremblay YDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, Briandet R, Dupuy B. A microbiota-generated bile salt induces biofilm formation in clostridium difficile. NPJ Biofilms Microbio. 2019;5(1):14. doi: 10.1038/s41522-019-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bechon N, Ghigo JM. Gut biofilms: bacteroides as model symbionts to study biofilm formation by intestinal anaerobes. FEMS Microbiol Rev. 2022;46(2). doi: 10.1093/femsre/fuab054. [DOI] [PubMed] [Google Scholar]
- 154.Michalak L, Gaby JC, Lagos L, La Rosa SL, Hvidsten TR, Tetard-Jones C, Willats WGT, Terrapon N, Lombard V, Henrissat B, et al. Microbiota-directed fibre activates both targeted and secondary metabolic shifts in the distal gut. Nat Commun. 2020;11(1):5773. doi: 10.1038/s41467-020-19585-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17(1):13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 156.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13(10):605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 2015;15(6):375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 158.Zavan L, Bitto N, Kaparakis-Liaskos MJI. History, vesicles dobm: bacterial membrane vesicles. Biogen Funct Applicat. 2020;1:1–21. [Google Scholar]
- 159.Dean SN, Leary DH, Sullivan CJ, Oh E, Walper SA. Isolation and characterization of lactobacillus-derived membrane vesicles. Sci Rep. 2019;9(1):877. doi: 10.1038/s41598-018-37120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dean SN, Rimmer MA, Turner KB, Phillips DA, Caruana JC, Hervey W, Leary DH, Walper SA. Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Front Microbiol. 2020;11:710. doi: 10.3389/fmicb.2020.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiol. 2012;158(Pt 11):2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 162.Elhenawy W, Debelyy MO, Feldman MF, Whiteley M, Greenberg EP. Preferential packing of acidic glycosidases and proteases into bacteroides outer membrane vesicles. mBio. 2014;5(2):e00909–00914. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host & Microbe. 2015;17(5):672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, Carding SR. Cephalosporinases associated with outer membrane vesicles released by bacteroides spp. protect gut pathogens and commensals against beta-lactam antibiotics. J Antimicrob Chemother. 2015;70(3):701–709. doi: 10.1093/jac/dku466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.James C, Dixon R, Talbot L, James SJ, Williams N, Onarinde BA. Assessing the impact of heat treatment of food on antimicrobial resistance genes and their potential uptake by other bacteria—a critical review. Antibiot (Basel). 2021;10(12):1440. doi: 10.3390/antibiotics10121440. [DOI] [PMC free article] [PubMed] [Google Scholar]


