Abstract
Marrow niches in osteosarcoma (OS) are a specialized microenvironment that is essential for the maintenance and regulation of OS cells. However, existing animal xenograft models are plagued by variability, complexity, and high cost. Herein, we used a decellularized osteosarcoma extracellular matrix (dOsEM) loaded with extracellular vesicles from human bone marrow-derived stem cells (hBMSC-EVs) and OS cells as a bioink to construct a micro-osteosarcoma (micro-OS) through 3D printing. The micro-OS was further combined with a microfluidic system to develop into an OS-on-a-chip (OOC) with a built-in recirculating perfusion system. The OOC system successfully integrated bone marrow niches, cell‒cell and cell–matrix crosstalk, and circulation, allowing a more accurate representation of OS characteristics in vivo. Moreover, the OOC system may serve as a valuable research platform for studying OS biological mechanisms compared with traditional xenograft models and is expected to enable precise and rapid evaluation and consequently more effective and comprehensive treatments for OS.
Keywords: 3D printing, Osteosarcoma-on-a-chip, Organ-on-a-chip, Bone marrow niches
Graphical abstract
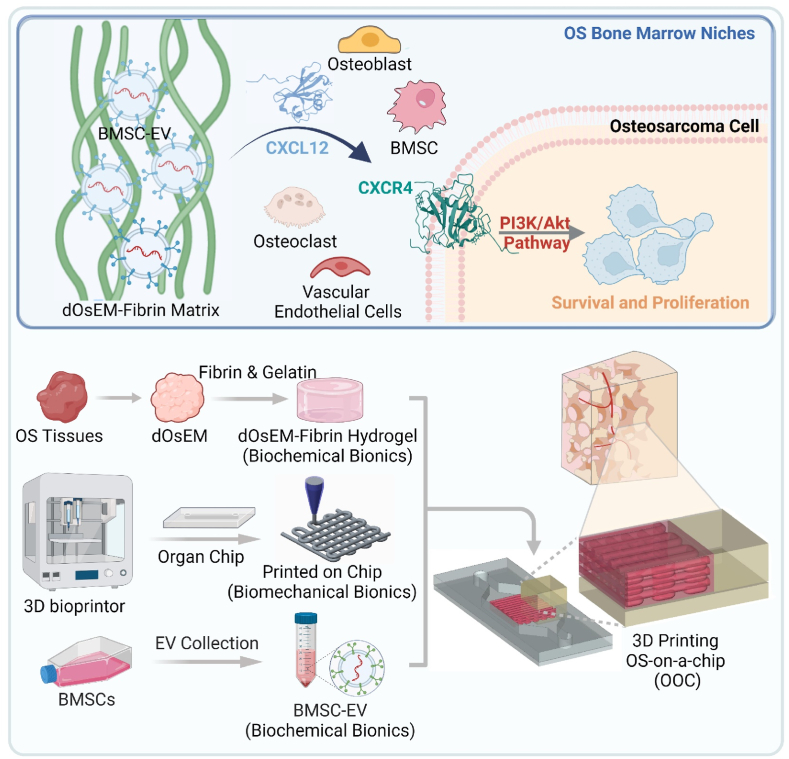
Schematic diagram showing the marrow-inspired OOC system could intimate bone marrow niches, cell-cell/cell-matrix crosstalk, and circulation, allowing for a more accurate portrayal of OS characteristics and drug responses in vivo.

Highlights
-
•
The significance of this study is demonstrated by the following aspects: (1) First, the decellularized osteosarcoma extracellular matrix (dOsEM) loaded with extracellular vesicles of bone marrow-derived stem cells (BMSC-EVs) was designed as an acellular bioink to construct a micro-osteosarcoma (micro-OS) for the expansion of biopsy samples and to maintain the biological features of OS tissue. (2) An OS-on-a-chip (OOC) system was established from the micro-OS combined with microfluidics, which imitated bone marrow niches, cell-cell and cell-matrix crosstalk, and circulation, allowing for a more accurate portrayal of OS characteristics in vivo. (3) The OOC system revealed that marrow niches regulated OS pathological processes and could serve as potential targets for OS treatment in the future.
1. Introduction
Osteosarcoma (OS), the most common primary malignant bone tumor, has an unsatisfactory prognosis due to its high aggressiveness and rapid metastasis. Existing research evaluating novel therapies and metastasis in OS research mostly involves 2D culture approaches and xenograft mouse models [1]. However, traditional 2D culture approaches do not accurately replicate the physicochemical characteristics of the ECM microenvironment of native cancer in vivo [2,3]. Thus, they do not conserve the original shape and polarization of cells and exhibit discrepant expression levels for key genes involved in angiogenesis, cell migration, and invasion. In contrast, xenograft mouse models (tumor-bearing models) involve the utilization of immune-deficient animals injected with human tumor cells (MG63 cell lines [4]), which have been shown to better mimic tumor growth and metastasis in vivo [5,6]. However, xenograft models can rarely completely recapitulate the formation of bone tumors in the same anatomical locations as in humans and with spontaneous development of metastases [1]. Xenograft models are also plagued by variability, complexity, and high cost. Therefore, neither type of model can be used for rapid and accurate simulation of the response of human tumors to drugs [7], and a more accurate and rapid model is needed for the study of OS stroma-cell interactions.
Tumor biomimetic systems, such as the many variations of three-dimensional (3D) tumor-on-a-chip models, are important platforms for studying tumor physiology and pathological functions in vitro. Our previous research showed that the chip model could combine the advantages of human cell culture models in vitro and rodent models in vivo to accurately simulate the characteristics of the tumor ECM [[8], [9], [10]], as well as the functions of tumor cell morphology, proliferation, drug metabolism, gene expression, and protein synthesis [9,11,12]. Moreover, it could reveal how specific components within the tissue and organ microenvironment influence cancer development and visualize how individual cancer cells behave over time when they interact locally with stromal cells [13]. In the case of OS, most tumor-on-a-chip studies have focused on the influence of molecular gradients on cell morphology or on the adhesive properties of OS cells under shear flow or at various temperatures and pH values [14,15]. In addition, due to current limitations of processing and biofabrication technologies, most existing tumor-on-a-chip models still cannot recapitulate the spatial structure of OS on a microscopic level or the cell distribution inside the OS matrix [16]. 3D printing is a technology that can introduce biomaterials to regulate cell functions by incorporating ECM ingredients, which facilitates the delivery of oxygen and nutrients in the OS matrix [17]. Therefore, it has great potential for constructing a particular arrangement and orientation of ECM constituents [18] on the tumor-on-a-chip to form an OS-specific microenvironment that supports OS cell adhesion and migration for further study of OS progression.
Recent studies have revealed that the stromal microenvironment of OS, which is composed of extracellular matrix (ECM) as well as cell secretions containing extracellular vesicles (EVs) and signaling molecules, is closely linked to OS occurrence and development [19]. These three components form a three-dimensional molecular network system that supports the physical structure, maintains the biochemical homeostasis of the tumor microenvironment [20], and regulates most tumor cell behaviors, including cancer cell invasion [21], proliferation, drug resistance [22,23], cancer stem cell maintenance [24], and immunosurveillance evasion [22,25,26]. Bone marrow-derived stem cells (hBMSCs), an essential component of the bone marrow microenvironment, are involved in the proliferation and migration of tumor cells [27], formation of the tumor microenvironment, and interaction with tumor cells [28]. Accumulating evidence shows that paracrine secretion of cytokines, such as IL-6, VEGF, and TNF-alpha, promotes the growth, metastasis, and immune escape of tumor cells [29]. As the main loading medium of secretory factors, EVs secreted by human bone marrow stem cells (hBMSC-EVs) are vesicles with a diameter of 40–100 nm that exist in the extracellular environment and contain multiple bioactive proteins, lipids, and nucleic acids [30]. In recent years, many studies have confirmed that BMSCs affect the occurrence and progression of OS [31]. Our previous studies indicated that EVs can mimic the effects of their parental cells of origin [32,33], induce the formation of blood vessels in tumors by regulating angiogenesis, stability, and maturation, and promote the proliferation of cells in tumor tissues [3,34,35]. Therefore, integrating the OS ECM with hBMSC-EVs could better simulate the stromal microenvironment of OS, facilitating the expansion of patient biopsy samples, the study of OS stroma–cell interactions, and the response of human tumors to drugs.
Here, we used a decellularized osteosarcoma extracellular matrix (dOsEM) with fibrin to construct a dOsEM-fibrin hydrogel loaded with hBMSC-EVs as an acellular bioink (dOsEM-EVs) to accurately replicate the physicochemical characteristics of the OS microenvironment. A 3D micro-osteosarcoma (micro-OS) was further constructed with dOsEM-EVs through 3D printing to recapitulate the spatial structure and cell distribution of OS. Then, the micro-OS combined with a microfluidic system was integrated into a multistage biomimetic osteosarcoma-on-a-chip (OOC) with a built-in recirculating perfusion system to test its drug screening potential. Our study established the feasibility of using a microfluidic OS chip model to precisely recapitulate the microenvironment of OS marrow niches, which facilitated comprehensive evaluations for developing more effective treatments for OS.
2. Results
2.1. Acquisition and characterization of the dOsEM
To replicate the high proliferation rate and metastatic characteristics of OS in vivo, we generated decellularized extracellular matrix from OS tissues (Fig. 1A, B). SEM results first demonstrated that dOsEM maintained the fibrous stromal composition of the OS tissue (Fig. 1C), which might drive from the collagen fibers. Then, quantitative analysis of the components inside dOsEM during decellularization showed that the cellular DNA and GAG contents were significantly reduced to 1.49 % ± 0.78 % (4.28 ± 2.20 ng/mg) and 18.55 % ± 1.34 %, respectively, while the content of the bioactive ingredient collagen was maintained at 118.65 % ± 1.36 % (Fig. 1D). To further quantitative analysis of other bioactive components, Mass spectrometry results showed that dOsEM was rich in ECM components such as collagen, fibrinogen, and laminin, which were found to promote cell adhesion and proliferation, as evidenced by Gene Ontology (GO) analysis (Fig. 1E and F). Besides, XPS results showed that the mineral components such as Ca and P could be detected inside the lyophilized dOsEM powder (Supplementary Fig. S1). These results showed that the mineral components could be maintained inside dOsEM component after decellularization process. However, some active components in the ECM (such as fibrinogen) were reduced from 14.645 % to 0.236 % during decellularization, which might derive from tissue mechanical disruption processes [36]. Therefore, to improve the biomimicry of the components, dOsEM-fibrin bioink was constructed by supplementing with fibrinogen and gelatin to further improve the stability of the structure (Fig. 1B).
Fig. 1.
Preparation and characterization of the dOsEM-fibrin hydrogel. (A) Schematic illustration of the fabrication of the dOsEM-fibrin hydrogel. (B) Preparation of the dOsEM-fibrin hydrogel. Scale bars represent 1 cm. (C) SEM identification of the microscopic topography within the dOsEM. Scale bars represent 200 μm. (D) Quantitative analysis of DNA, collagen, and GAG components of the dOsEM and OS tissues (n = 4). (E) Mass spectroscopy detection of the components of dOsEM. (F) GO analysis of the dOsEM proteins detected using mass spectrometry. (G) Rheological characterization of the dOsEM-fibrin hydrogels. Dependence of the storage modulus (G′) and loss modulus (G″) on the shear strain of dOsEM-fibrin hydrogel materials with gelatin concentrations from 0 % to 3 %. (H) Dependence of the loss factor on the shear strain of dOsEM-fibrin hydrogel materials with gelatin concentrations from 0 % to 3 % (n = 3). (I) MTT assay of cell proliferation inside dOsEM-fibrin hydrogels in vitro (n = 8). (J) Cell adhesion and viability of the OS cell line MG63 inside the dOsEM-fibrin hydrogel. Scale bars represent 100 μm. (K) RNA-seq combined with KEGG analysis for cell proliferation, cell adhesion, and matrix–receptor interactions inside the dOsEM-fibrin hydrogel. Analysis of variance (ANOVA) was performed for all parameters. Newman‒Keuls comparison tests were performed after ANOVA. Mean ± SD are presented. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
2.2. Preparation and biological characterization of the dOsEM-fibrin hydrogel for 3D printing
Rheological properties are crucial for material processing. The loss factor is an important indicator of viscoelastic self-supporting properties and is defined as the ratio between the loss modulus (G″) and the storage modulus (G’). A value less than 1 means predominantly elastic behavior that enables self-support in a definite shape without easy deformation. Our results showed that with a gelatin concentration from 2 % to 3 % (w/w), the dOsEM-fibrin hydrogel behaved as a viscoelastic solid that was self-supporting before 3D printing (Fig. 1G and H, Supplementary Fig. S2), making it an ideal bioink in terms of rheological properties.
Subsequently, the proliferation activity of the OS cell line MG63 inside dOsEM-fibrin was compared with that in four other commercial hydrogels (GelMA hydrogel [37], hyaluronan/Gelin/PEGDA hydrogel [38], collagen hydrogel and fibrin hydrogel). The results showed that the prepared dOsEM-fibrin hydrogel enabled the highest proliferation in vitro, which might derive from the fact that the bioactive macromolecules and small-molecule components in dOsEM-fibrin are partly retained, promoting higher proliferation of OS cells than other commercial hydrogels, which is consistent with the results of related research [39]. (Fig. 1I). Confocal imaging of cytoskeleton staining by phalloidin showed that OS cells inside the dOsEM-fibrin group displayed stretched cytoskeletons with extended cytoplasmic protrusions and exhibited strong motility and higher proliferation activity in the 3D microenvironment than the Fibrin hydrogel (Fig. 1J). As expected, RNA-seq analysis showed that the dOsEM-fibrin hydrogel significantly promoted gene expression corresponding to the cell cycle, DNA replication, cell adhesion, and ECM-receptor interaction (Fig. 1K). Taken together, these results demonstrated that dOsEM-fibrin is an ideal bioink for cell proliferation, cell adhesion, and matrix–receptor interactions in OS cells.
2.3. hBMSC-EVs promote the proliferation and migration of OS cells
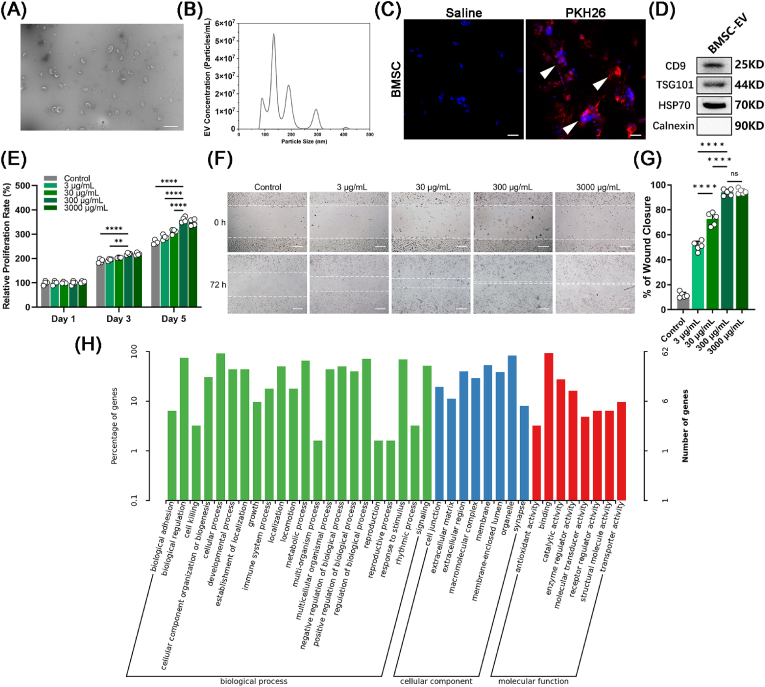
After characterizing the morphology of the hBMSCs (Supplementary Fig. S3), hBMSC-EVs were collected. The concentration of hBMSC-EVs was 1.15 ± 0.07 mg/mL (3860 particles/cell). TEM showed that hBMSC-EVs had a typical round vesicular morphology [40] (Fig. 2A). The size distribution detected by nanoparticle tracking analysis (NTA) showed that the average diameter of the hBMSC-EVs was 133.3 ± 4.5 nm (Fig. 2B). Staining hBMSC-EVs with PKH26 [41] revealed that they fused with OS cell membranes, which facilitated the release of small molecules into OS cells [42] (Fig. 2C). Western blot analysis verified the presence of CD9, TSG101, and HSP70 (Fig. 2D), which are three typical markers for hBMSC-EVs, as well as the absence of calnexin, which is a negative marker for hBMSC-EVs but a positive marker for hBMSCs [43].
Fig. 2.
Characterization of hBMSCs and hBMSC-EVs. (A) TEM topography of hBMSC-EVs. Scale bars represent 500 nm. (B) Particle size distribution of hBMSC-EVs using NTA (n = 3). (C) Cellular phagocytosis assay of hBMSC-EVs. Scale bars represent 50 μm. Positive staining is indicated by a white arrow. (D) Western blot analysis of hBMSC-EV-specific markers (CD9, TSG101, and HSP70). (E) hBMSC-EVs promoted the proliferation of MG63 cells. MTT assays measured the proliferation effects of different concentrations of hBMSC-EVs, 3 μg/mL, 30 μg/mL, and 300 μg/mL (n = 4). Scale bars represent 200 μm. (F, G) A scratch test was used to measure cell migration with different concentrations of hBMSC-EVs, 3 μg/mL, 30 μg/mL, and 300 μg/mL (n = 6). (H) The biological process, cellular component, and molecular function analysis showed that the hBMSC-EVs promoted cell proliferation, cell adhesion, and cell–matrix interactions. Analysis of variance (ANOVA) was performed for all parameters. Newman‒Keuls comparison tests were performed after ANOVA. The mean ± SD are presented. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
Then, we measured the proliferation under different concentrations of hBMSC-EVs from 3 μg/mL to 300 μg/mL hBMSC-EVs at 300 μg/mL showed the highest facilitation of OS cell growth (Fig. 2E). The scratch assay verified that 300 μg/mL hBMSC-EVs also showed the highest migration rate (Fig. 2F and G). The differentially expressed genes after hBMSC-EV treatment were examined using GO and KEGG analysis. The results showed that the hBMSC-EVs significantly promoted cell proliferation, cell adhesion, and matrix–receptor interactions (Fig. 2H, Supplementary Fig. S4). Taken together, these results showed that hBMSC-EVs promoted OS cell proliferation, cell adhesion, and cell–matrix interactions.
2.4. 3D-printed micro-OS models simulate OS marrow niches to maintain OS cell viability and characteristics in vitro
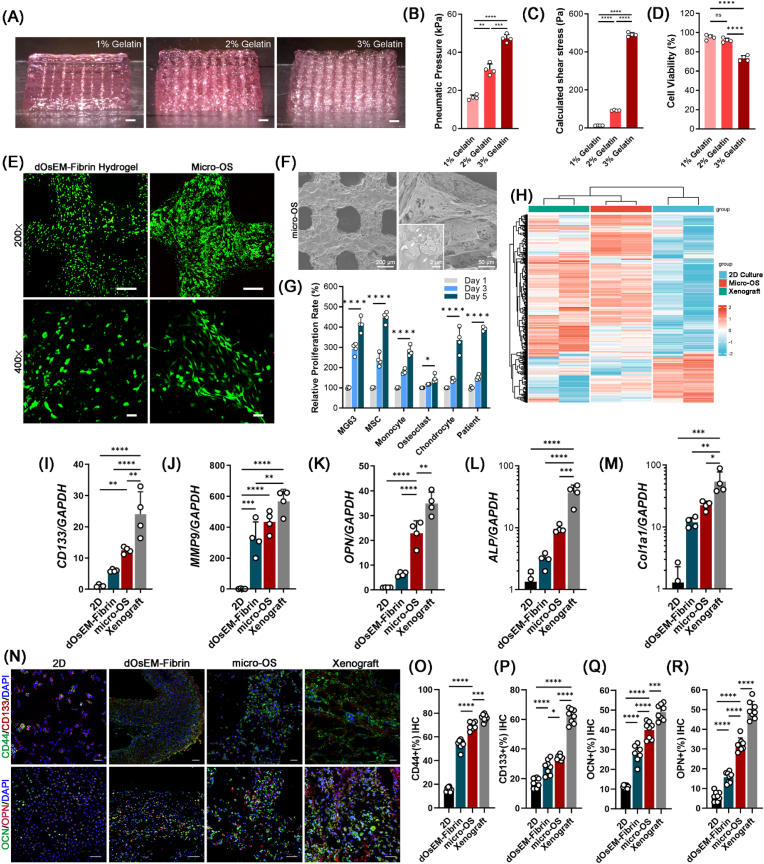
We then used dOsEM-fibrin mixed with hBMSC-EVs as an acellular bioink to construct micro-OS tissue with MG63 cells. The results showed that the molding properties of the printed micro-OS improved as the gelatin concentration was increased (Fig. 3A). However, the pneumatic pressure and the calculated shear stress increased dramatically (Fig. 3B and C), which caused more cell death [44] (Fig. 3D). Therefore, 2 % (w/w) gelatin was the best option for good formability and low shear damage. We further print the osteosarcoma-specific structures with 2 % gelatin based on the Computed Tomography (CT) pictures of patient's OS tissue (Supplementary Fig. S5). By comparing the Young's Modulus between OS tissue and Micro-OS, the results showed that the modulus (stiffness) of the prepared micro-OS tissue was closed to the OS ECM in vivo (Supplementary Fig. S6), which illustrate physiological relevancy between the 3D printed micro-OS and the OS tissues from patients.
Fig. 3.
The fabrication and characterization of the micro-OS. (A) Side view of the 3D-printed micro-OS scaffold with different concentrations of gelatin. (B) Printing pneumatic pressure and calculated shear stress (C) of different concentrations of gelatin (n = 4). (D) The viability of OS cells in the 3D-bioprinted micro-OS after extrusion through the nozzle. (E) Live/dead staining of OS cells in the 3D-printed dOsEM-fibrin hydrogel and 3D-bioprinted micro-OS under 200 × and 400 × magnifications. Cells were stained with CaAM (green) and PI (red). Scale bars represent 100 μm (upper) and 40 μm (lower). (F) SEM images of the 3D micro-OS and the hBMSC-EVs inside (white arrow). Scale bars represent 200 μm (left) and 20 μm (right). (G) Proliferation of MG63 cells, MSCs, monocytes, osteoclasts, chondrocytes and patient-derived OS cells inside the micro-OS (n = 4). (H) Expression profiling of genes with RNA-seq analysis for the 2D culture, micro-OS, and xenograft model. (I) MG63 expression of the cell proliferation-related genes CD133, MMP9 (J), and OPN (K) was enhanced inside the micro-OS compared to the 2D culture and xenograft model. (K) Osteo-related gene expression of OPN, ALP (L) and COL-1 (M) in the micro-OS (n = 4). (N) Immunofluorescence detection and quantification of the expression of CD44 (O), CD133 (P), OCN (Q) and OPN (R) in MG63 cells inside the micro-OS (n = 8). Scale bars represent 200 μm. Analysis of variance (ANOVA) was performed for all parameters. Newman‒Keuls comparison tests were performed after ANOVA. Mean ± SD are presented. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
Live/dead staining showed that the 3D-printed micro-OS model had better cell adhesion viability and morphology than the 3D-printed dOsEM-fibrin hydrogel (Fig. 3E). The SEM results verified the presence of hBMSC-EVs (Fig. 3F). In addition to OS cell lines (MG63 cells), micro-OS model significantly maintained the proliferation of different cells in the marrow niche, including MSCs, monocytes, osteoclasts, fibroblasts, and chondrocytes (Fig. 3G). Most importantly, micro-OS model also supported the proliferation of patient-derived OS cells (Fig. 3G). As the modulus (stiffness) has been proved to be vital for OS cells attachment and proliferation capabilities [20,21], the similarities in modulus of the prepared micro-OS tissue and the OS ECM in vivo might explain the similarities in cell proliferation behavior within these two groups (Supplementary Fig. S6).
To further examine the differences in gene expression in micro-OS and in the typical 3D model in vivo (xenograft model), we performed hierarchical clustering analysis of metabolism-related gene expression profiles. The results showed that cells inside the micro-OS clustered in a manner more similar to cells in vivo conditions (xenograft model) than cells in 2D culture (Fig. 3H). The gene expression of cancer-specific markers (e.g., CD133 and MMP9) and osteo-related genes (e.g., OPN, ALP and COL-1a1) in the micro-OS was elevated (Fig. 3I–M). Immunohistochemical analysis of CD44, CD133, OPN and OCN (Fig. 3N) further verified that the micro-OS mimicked the tumor microenvironment in vitro more accurately than 2D culture or culture in a 3D-printed dOsEM-fibrin hydrogel (Fig. 3O–R). These results suggested that the micro-OS could support the viability and maintain the characteristics of OS cells, comparing with the 2D culture approaches and xenograft mouse models.
2.5. The marrow inspired OOC system mimicked OS marrow niches and maintained the high aggressiveness and rapid metastasis of OS tissues in vitro
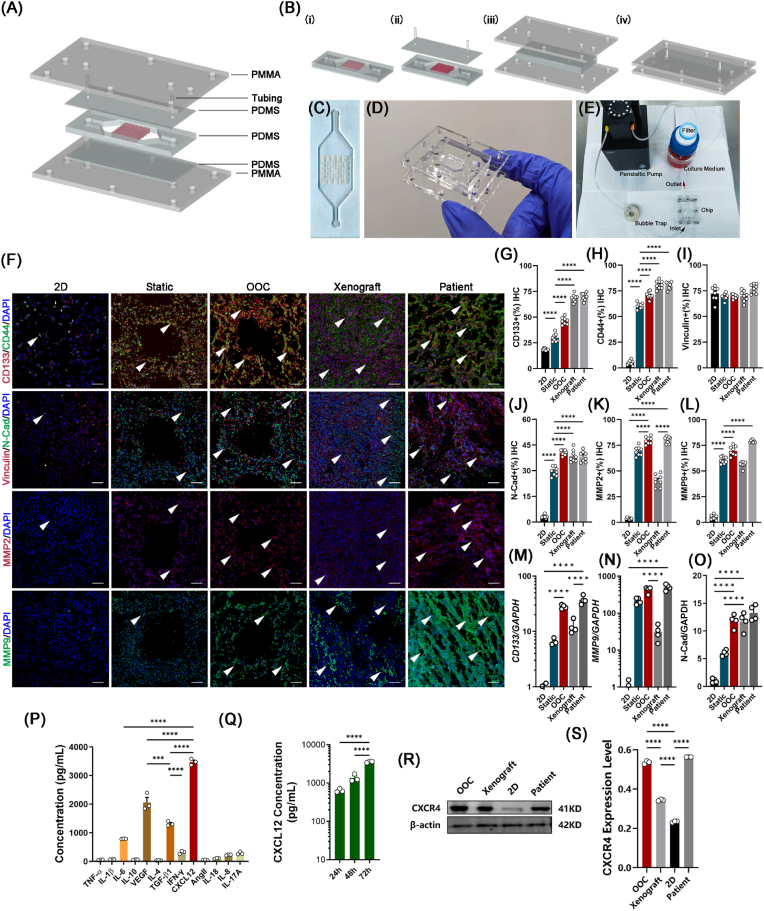
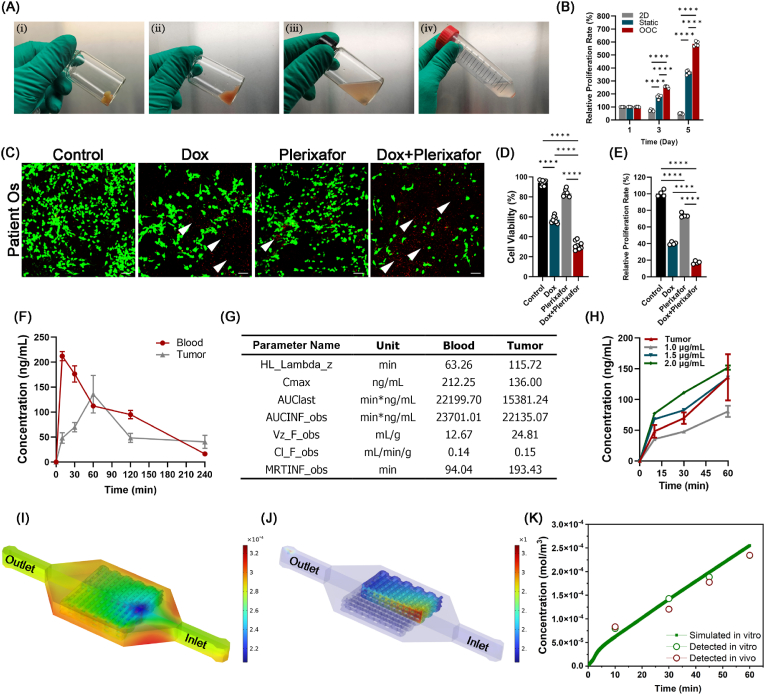
An OOC system was then constructed that integrated the micro-OS with a microfluidic platform to simulate the diffusion behavior of drugs within the circulatory system inside the bone marrow niches (Fig. 4A–E). IHC staining for the cancer stemness marker CD133 in the OOC system was 46.37 % ± 3.40 %, which was significantly higher than that in the 2D and static models (Fig. 4F and G). The relative expression level of CD44 was also significantly elevated in the OOC system compared to the 2D and static (without perfusion) models (Fig. 4H and I). Positive staining for N-cadherin, a typical marker for metastasis, increased from 3.31 % ± 0.33 % in the 2D culture to 40.63 % ± 0.53 % in the OOC system, which was higher than that in the static model (Fig. 4J). In the OOC system, the secretion of MMP2 and MMP9, which contribute to OS invasion and metastasis, was significantly increased (MMP2: 79.89 % ± 1.22 % and MMP9: 69.69 % ± 4.57 %) (Fig. 4K and L). Then, we further compared the gene expression of CD133, MMP9, and N-cadherin among the 2D, static, OOC, and xenograft models and patient OS tissue, and the results confirmed that the aggressive and metastatic characteristics of OS were well maintained in the OOC system (Fig. 4M − O). Taken together, these results illustrated that the OOC system mimicked the OS marrow niches to maintain high OS aggressiveness and rapid metastasis and might serve as an ideal tool for individualized drug screening.
Fig. 4.
Fabrication of the osteosarcoma-on-a-chip (OOC) system and immunofluorescence detection of metastatic markers of tumor cells inside the OOC system. (A) Schematic illustration of the 3D-printed OOC model. (B) Steps for in situ fabrication of a 3D-printed OOC model. (C–E) Experimental setup of the OOC model. The chip was connected in a closed loop to a reservoir with medium and drugs passing through a peristaltic pump. (F) Immunofluorescence detection and quantification of the expression levels of CD133 (G) and CD44 (H), vinculin (I), N-cadherin (J), MMP2 (K) and MMP9 (L) inside the 2D, static, OOC, and mouse xenograft models and patient OS tissue (n = 8). Scale bars represent 200 μm. Positive staining is indicated by a white arrow. (M) RT‒qPCR measurement of the expression levels of CD133, MMP2 (N), and N-cadherin (O) in MG63 cells inside 2D, static, OOC, and mouse xenograft models and patient OS tissue (n = 4). Expression was normalized to that of the 2D culture conditions. (P) Expression of cytokeratin concentrations inside hBMSC-EVs (n = 3). (Q) Concentration of CXCL12 inside hBMSC-EVs at 24, 48, and 72 h (n = 3). (R) Expression and quantification (S) of CXCL12 with different volumes (30 μL and 300 μL) of hBMSC-EVs. CXCL12 protein was used as the positive control. (T) Expression and quantification (U) of the CXCL12-specific receptor CXCR4 in 2D, static, and OOC models and patient OS tissue. (n = 3). Data are presented as the means ± SDs (n = 4). Analysis of variance (ANOVA) was performed for all parameters. Newman‒Keuls comparison tests were performed after ANOVA. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
2.6. The OOC system exhibits high OS aggressiveness and rapid metastasis through activation of the CXCL12/CXCR4 signaling axis
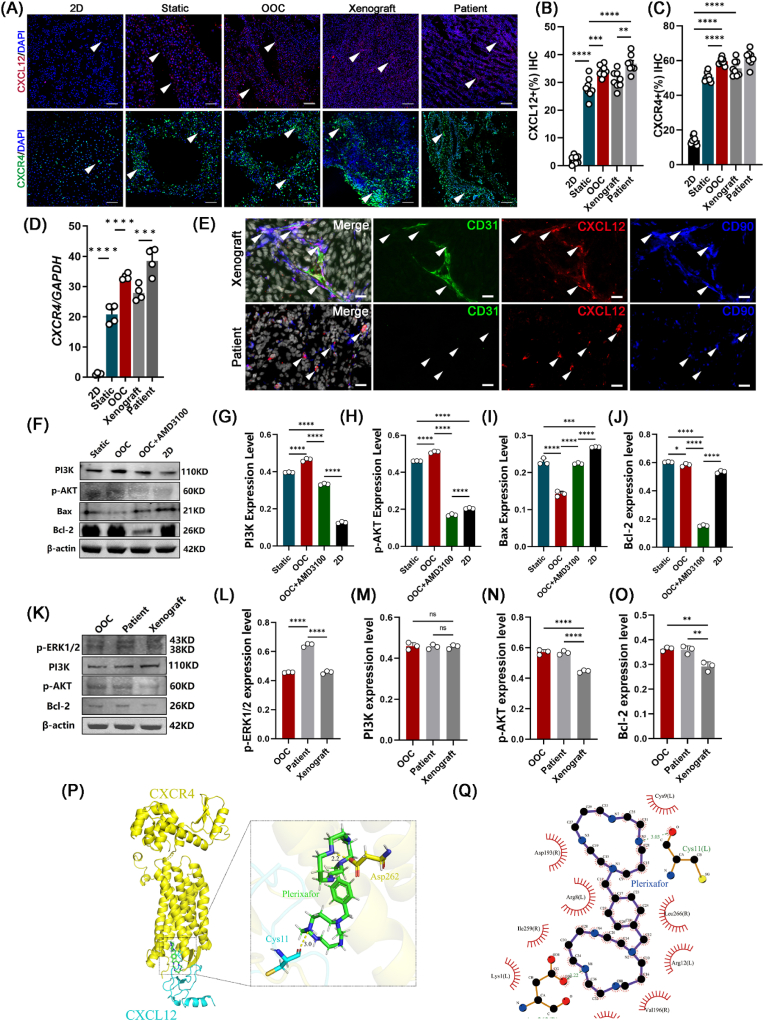
An in vivo biomimetic platform could provide real-time, accurate, and individualized results for OS drug screening. We next explored the mechanisms by which the artificial OS marrow niches (the OOC system) maintained the high aggressiveness and rapid metastasis of OS cells. Previous results (Supplementary Fig. S4) showed that the ECM and ECM-receptor interactions significantly affected the characteristics and maintenance of OS cells. Thus, we performed a STRING analysis to determine the key secreted protein candidates inside the OOC system. Cytokine-related proteins were the key clustered molecules (Supplementary Fig. S7). ELISA results further demonstrated that CXCL12 accounted for a large proportion of the hBMSC-EV-secreted proteins (approximately 3478.473 ± 86.979 ng/mL) (Fig. 4P-Q). Apart from CXCL12, its receptor CXCR4 showed greater enhancement in the dOsEM groups than in the 2D or static group (Fig. 4R and S). Immunohistochemical analysis (Fig. 5A–C) and gene expression analysis (Fig. 5D) showed that the expression of CXCR4 and CXCL12 in the OOC system was more similar to that in xenograft and patient OS tissues. Moreover, our xenograft model and patient OS tissue model also verified the existence of CXCL12, which was mainly secreted by CD90-positive mesenchymal stem cells in vivo, rather than vascular endothelial cells or tumor cells (Fig. 5E), which was consistent with the results of a previous study [45] and further clarified that adding hBMSC-EVs to the OOC system in vitro could partially restore the OS bone marrow niches and the related CXCL12/CXCR4 interactions in vivo.
Fig. 5.
Activation of the CXCL12/CXCR4 signaling pathway increased proliferation characteristics in the OOC system. (A) Immunofluorescence detection and quantification of the expression levels of CXCL12 (B) and CXCR4 (C) in the 2D culture, static, OOC, and mouse xenograft models and patient OS tissue. (n = 8). Scale bars represent 200 μm. Positive staining is marked as a white arrow. (D) RT‒qPCR detection of the expression levels of CXCR4 in MG63 cells inside the 2D, static, OOC, and mouse xenograft models and patient OS tissue (n = 4). (E) Immunofluorescence detection of the localization of CXCL12 (red), CD90 (blue), and CD31 (green) inside the mouse xenograft model and patient-derived OS tissues. Scale bars represent 10 μm. (F) Protein expression levels and quantification of PI3K (G), p-AKT (H), Bax (I), and Bcl-2 (J) for the static model, OOC, and OOC with CXCR4 inhibitor (AMD3100) and 2D culture (n = 3). (K) Protein expression levels and quantification of p-ERK1/2 (L), PI3K (M), p-AKT (N), and Bcl-2 (O) in the OOC model, patient OS tissue, and xenograft model (n = 3). (P) Construction of the CXCR4–CXCL12 complex by protein‒protein docking. (Q) 3D imaging and (R) 2D imaging of molecular docking for the plerixafor and CXCR4–CXCL12 complex. The carboxyl hydroxyl group O–H of the Asp262 residue (yellow) of the CXCR4 protein formed a hydrogen bond of 2.2 Å with the N of plerixafor (green). The main chain carboxyl group O of the Cys11 residue (blue) of the CXCL12 protein formed a hydrogen bond with a length of 3.0 Å with an N–H group of plerixafor (green). Data are presented as the means ± SDs. Analysis of variance (ANOVA) was performed for all parameters, followed by a Newman‒Keuls comparison test. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
We then verified whether the restoration of CXCL12/CXCR4 interactions is closely related to cell proliferation enhancement inside our OOC system. Previous results showed that the classic proliferation regulatory pathway PI3K/AKT was upregulated in our OOC system (Supplementary Fig. S8). Western blotting also verified the increased protein expression of PI3K and p-AKT, which was consistent with the RNA-seq results (Fig. 5F-O). The addition of AMD3100 (also known as plerixafor), a small-molecule inhibitor that specifically inhibits CXCR4 receptors, offset the regulatory effects of the OOC system by decreasing the expression of PI3K and p-AKT proteins (Fig. 5F). We then determined the inhibitory mechanism of AMD3100 on the CXCR4 signaling axis through protein docking and molecular docking, and the results showed an inhibitory effect of AMD3100 on the CXCL12/CXCR4 axis that was exerted by targeting the Asp262 residue of CXCR4 and the Cys11 residue of CXCL12 (Fig. 5P, Q, Supplementary Figs. S9 and S10).
Taken together, these results indicated that the OOC system simulated marrow niches and maintained PI3K/AKT-mediated proliferation and metastasis in OS cells by sustaining the CXCL12/CXCR4 axis.
2.7. Evaluation of therapeutic effects in the OOC system
Because the OOC system exhibits high OS aggressiveness and rapid metastasis through the CXCL12/CXCR4 axis, we then evaluated the potential of the OOC system in drug screening and compared it with a xenograft model. DOX is the first-line chemotherapy drug for OS. We combined DOX with AMD3100 (plerixafor) in our OOC system to verify the efficiency of comprehensive therapy. The primary OS specimens were freshly isolated from the OS patient (Fig. 6A). The results showed that the proliferation of patient-derived OS cells in the OOC group was greater than that in the 2D and static groups (Fig. 6B), which is consistent with previous research results [10]. The results of the viability test preliminarily revealed that although the killing effect of plerixafor was not as strong as that of DOX, the combination of plerixafor (40 μM) and DOX (0.8 μM) significantly increased the lethal effect of DOX on patient-derived OS cells. The cell viability decreased from 57.07 % ± 3.39 % (for DOX alone) to 31.52 % ± 4.23 % (for the combination group) (Fig. 6C and D), and the cell proliferation measurement through the CCK8 assay showed similar results (Fig. 6E). Then, we used the OOC system to compare the pharmacokinetic and pharmacodynamic properties of different concentrations of DOX with the xenograft model in vivo (Fig. 6F and G). We found that 1.5 μg/mL DOX in the OOC system more closely simulated the concentration changes in vivo (Fig. 6H). The model prediction of the average volume of the OOC system successfully simulated the concentration changes in the in vivo model (Fig. 6I–K). Taken together, these results suggested that the OOC system is an effective model for drug screening and toxicity prediction.
Fig. 6.
Evaluation of the drug screening potential of the OOC system. (A) Primary OS specimens were freshly isolated from the OS patient (i), and after being minced with scissors (ii), OS tissues were digested with tissue dissociation solution under magnetic stirring (iii). The digested cells were then separated from the digestion solution by centrifugation at 1300 rpm for 10 min (iv). (B) Proliferation of patient-derived OS cells inside the 2D, static and OOC systems. (C) The combination of plerixafor and DOX significantly increased the killing effect of DOX on OS cell viability (D) and proliferation (E). Scale bars represent 50 μm. PI staining of dead cells is indicated by a white arrow. (F, G) The pharmacokinetic and pharmacodynamic properties of DOX in OS tissues (n = 4). HL_Lambda_z: Terminal half-life = ln (2)/Lambda Z; Cmax: Maximum observed concentration; AUClast: Area under the curve from the time of dosing to the time of the last measurable (positive) concentration (Tlast); AUCINF_obs: AUC from time of dosing extrapolated to infinity, based on the last observed concentration (_obs) or last predicted concentration; Vz_F_obs: Volume of distribution based on the terminal phase; Cl_F_obs: Total body clearance for extravascular administration; MRTINF_obs: Mean residence time (MRT) extrapolated to infinity. (H) Three concentrations of DOX in the medium (n = 4). (I, J) Simulation results of DOX distribution in the 3D-printed OOC model for 60 min. (K) Model prediction of the average volume of the 3D-printed OOC model compared to the in vitro and in vivo models. Data are means ± SDs for n = 4 replicates. Analysis of variance (ANOVA) was performed for all parameters, followed by a Newman‒Keuls comparison test. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
2.8. The xenograft model verified the therapeutic effect of the CXCR4 inhibitor plerixafor in the OOC system
Based on the abovementioned results, we further verified the effectiveness of the CXCR4 inhibitor plerixafor combined with traditional combination therapy (doxorubicin and cisplatin [46,47]) (Fig. 7A). The size of the tumor in nude mice decreased significantly in the APA group (Fig. 7B) compared to that in mice treated with traditional chemotherapy drugs (control: PBS; AP: doxorubicin and cisplatin; APA: AP and AMD3100). The weight changes of the mice showed no significant difference among the three groups (Fig. 7C and D). Immunofluorescence results further verified that CXCR4 expression was inhibited (Fig. 7E). Moreover, decreases were observed in the expression of tumor stem cell markers (CD44 and CD133) and tumor metastasis markers, such as N-cadherin and EpCAM for EMT processes and MMP2 and MMP9 for matrix degradation. (Fig. 7F–M). Caspase-3 staining showed significantly increased expression from 36.66 % ± 0.21 % to 84.52 % ± 0.76 % (Fig. 7N, P), and apoptosis increased from 23.69 % ± 0.63 % to 46.56 % ± 2.00 % (Fig. 7O, Q). Taken together, these results verified that our OOC system simulated the therapeutic effect of drugs in vivo and that plerixafor could be a potential targeted treatment for OS in the future.
Fig. 7.
Therapeutic effect of the CXCR4 inhibitor AMD3100 combined with the chemotherapy drug DOX on OS in vivo. (A) Schematic diagram of the administration of saline, AP, and APA in nude mice. (B) Effects of DOX and CXCR4 on MG63 cell tumor growth in vivo. (C) Tumor volume growth curves in the control, AP, and APA groups (n = 6). (D) Weight changes of mice in the control, AP, and APA groups (n = 6). (E) Immunofluorescence detection and quantification of the expression of CXCR4 (F), CD133 (G), CD44 (H), E-cadherin (I), MMP2 (J), EpCAM (K), MMP9 (L), and N-cadherin (M) in MG63 cells in the control, AP, and APA groups. Scale bars represent 200 μm. Positive staining is indicated by white arrows. (N–Q) Immunofluorescence detection and quantification of the expression levels of caspase-3 and TUNEL staining for the control, AP, and APA groups (AP: doxorubicin and cisplatin; APA: AP and AMD3100). Scale bars represent 200 μm. (n = 5). Data are presented as the means ± SDs. Analysis of variance (ANOVA) was performed for all parameters, followed by a Newman‒Keuls comparison test. *, **, ***, and **** indicate p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
In summary, the constructed bone marrow-inspired OOC system simulated marrow niches to support the high aggressiveness and metastasis of OS cells and can be used as a potential drug screening platform for OS in the future.
3. Discussion
Tissue-engineered 3D OS models have demonstrated great potential for studying the mechanism of OS cell–matrix interactions and assessing drug efficacy by incorporating key features of OS niches [48]. As the key component of OS niches, the ECM, extracellular vesicles and signaling molecules work together to constitute a three-dimensional molecular network system [20], which provides physical support for the structures and maintenance of biochemical functions for OS cells, such as cell invasion, migration and proliferation [21]. To imitate the actual microenvironment in vivo, and further determine its role in modulating OS progression and drug responses, it is necessary to construct an OS model with controllable matrix stiffness, cell‒cell interactions, cell–matrix interactions and a perfusion system.
Herein, this study created an acellular OS marrow niche (the OOC) system by combining dOsEM with marrow-specific hBMSC-EVs in a microfluidic recirculation perfusion system. The dOsEM was used as a soft material for modeling OS in 3D and maintained the biochemical properties of the bone tissue and the spatial patterning of the matrix during OS invasion. Our results first show that the amplification efficiency of dOsEM is significantly better than that of other commercial hydrogels (Fig. 1I). In addition, dOsEM-EVs in OOC have good expansion efficiency and could maintain important characteristics of the OS microenvironment (Fig. 2, Fig. 3). We speculate that the promotion of OS cell proliferation by the OOC system may be attributable to three reasons. (1) The 3D molecular network of dOsEM partially maintains the physical structure and biochemical homeostasis of the OS microenvironment after decellularization treatment. (2) The hBMSC-EVs provide a growth factor microenvironment, possibly derived from bioactive small-molecule contents. (3) The softness of the dOsEM in our OOC may also be vital for the maintenance of cancer stem cells and the related epithelial-mesenchymal transition (EMT) markers for osteosarcoma, such as CD133, CD44 and N-cadherin (Fig. 3I, N; Fig. 4F, M), which is consistent with previous research findings [49].
In addition, 3D printing technology integrated with the OOC perfusion system, as an efficient method to construct porous scaffolds with intricate microstructures, has been proven to be vital in controlling the oxygen and nutrient distribution to alleviate hypoxia [9,10,50] throughout the constructed OS tissue (Fig. 3E), which is indispensable in constructing high-viability tissues for drug screening applications [51,52]. Vasculature, another key component affecting tumor progression and treatment response, has also been proven to be necessary to provide sufficient oxygen and nutrients to cells within tissues above the millimeter scale and to convey circulating tumor cells (CTCs) outward for cancer metastasis [12,25,44]. Accordingly, future work will integrate the vasculature networks of 3D-printed organ-on-chip models into a vascularized multiorgan-on-chip model to facilitate preclinical pharmaceutical investigation and basic research on cancer biology.
Moreover, hBMSC-EVs, a human-derived biological component rich in bone marrow-specific secreted factors (such as CXCL12, IL-6, VEGF, and TGF-β), could mimic multiple biochemical components in the microenvironment (Fig. 4P). hBMSC-EVs have also been found to produce a sustained release of these secreted factors [53], facilitating long-lasting nourishment for OS cells, which is superior to simply adding these factors to the hydrogel. In addition, compared with the recent results of directly adding stem cells, this improved method significantly reduced stem cell contamination for future drug screening processes. Therefore, hBMSC-EVs might be used as an optional bone marrow replacement for human-derived hBMSCs inside the OOC system, and hBMSC-EVs could be used as effective additives for tissue amplification in vitro.
To further elucidate the specific regulatory mechanism of hBMSC-EVs on OS cells, we used RNA-seq and mass spectrometry to explore the possible crosstalk between BMCS-EVs and OS. The results showed that cytokine‒cytokine receptor interactions, especially CXCR4–CXCL12 interactions, played a vital role in their regulatory effect. CXC motif chemokine ligand 12 (CXCL12; SDF-1) is a chemokine of the CXC family, is often expressed in bone marrow stem cells, and interacts with CXC motif chemokine receptor 4 (CXCR4) [54]. CXCR4 has been shown to be expressed in several cancers, including breast, renal, prostate, and gastric cancers [55,56]. Studies have shown that mesenchymal stem cells promote the proliferation of OS cells through paracrine action and regulate the downstream pro-proliferation pathway by directly secreting CXCL12 [57]. CXCL12 has also been found to play a critical role in OS metastasis [58]. The expression of CXCL12 might be stimulated by HIF-1α during OS growth [59,60], which could further promote OS migration, angiogenesis, and stemness maintenance (e.g., CD133 and CD44) at a high cell density. As expected, our results showed that hBMSC-EVs upregulated the CXCL12/CXCR4 axis and shared a similar role in patient OS proliferation and migration (Fig. 5). hBMSC-EV-enriched secretory components (such as CXCL12) might have a strong synergistic effect with matrix stiffness (modulus) and bioactive ingredients remaining in dOsEM-fibrin after decellularization treatment (such as heparan sulfate [61] and hyaluronan [62]) for regulating cell behavior [63], further validating the necessity of this model for elucidating the pathological progression of OS. This is also reflected by the elevated expression of CXCR4 in our OOC system compared with other models (Fig. 5C and D).
AMD3100 (also known as plerixafor), a CXCL12 and CXCR4 inhibitor, has been approved by the FDA for patients with lymphoma and multiple myeloma [64]. However, its potential applications and the specific mechanism by which it regulates CXCR4 remain unclear. We found that AMD3100 in the OOC system impaired PI3K/AKT-mediated cell proliferation and migration (Figs. 5F and 6C), which was consistent with previous research results [65]. Molecular docking showed that the mechanism might drive competitive inhibition effects between the binding domain of CXCL12 and CXCR4 at the Asp262 residue in CXCR4 and the Cys11 residue in CXCL12 (Fig. 5Q–S). These results showed that CXCL12 may be a major factor in regulating OS proliferation and that PI3K/AKT could be the key downstream pathway under CXCL12/CXCR4 axis activation after AMD3100 treatment, which could regulate OS growth, metastasis, and drug resistance [66,67]. Further work would include constructing a lentivirus delivery system via CRISPR/Cas9 RNP to specifically knock out CXCR4 and further verify its killing effect on OS. Further research will explore the specific functions and mechanisms of other bioactive ingredients in BMSC-EVs (such as DNA/miRNA/lncRNA) in regulating OS cell proliferation and metastasis.
4. Conclusions
An acellular bioink consisting of dOsEM loaded with hBMSC-EVs was used to construct a micro-OS. By integration with a microfluidic system, the micro-OS was further developed into an OOC system, which imitated bone marrow niches, cell‒cell and cell–matrix crosstalk, and circulation, allowing a more accurate representation of OS characteristics in vivo. We confirmed that the marrow niche-inspired OOC system regulated multiple OS pathological processes via the CXCL12/CXCR4/PI3K/AKT pathway and that plerixafor, by targeting CXCR4, has potential as a targeted treatment for OS. The OOC system could also be used as a drug screening platform for OS to study the therapeutic effects of systemic therapies to design individualized comprehensive treatments in the future (Fig. 8).
Fig. 8.
Schematic diagram showing the marrow-inspired OOC system imitating bone marrow niches, cell‒cell and cell–matrix crosstalk, and circulation, allowing a more accurate representation of OS characteristics and drug responses in vivo.
5. Materials and methods
5.1. Preparation and characterization of the 3D-printed dOsEM-fibrin scaffold
5.1.1. Extraction and characterization of hBMSC-EVs
hBMSCs were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Then, P2 and P3 hBMSCs were used to extract hBMSC-EVs. When the cell confluency reached approximately 70 %, the cell culture medium was replaced with serum-free medium. The cell culture supernatant was collected after culturing for 72 h. Cell impurities were removed by centrifugation at 1200 rpm, and then ultracentrifugation was performed at 100,000×g and 4 °C for 90 min. The supernatant was hen discarded and resuspended in 0.5 mL of Dulbecco's phosphate-buffered saline (DPBS) to obtain the hBMSC-EVs. The concentration of hBMSC-EVs was determined using a BCA kit (Thermo Fisher, MA, USA).
The morphology of hBMSC-EVs was observed using transmission electron microscopy (TEM, TecnaiTM G2 Spirit BIOTWIN). The particle size distribution of hBMSC-EVs was observed using nanoparticle tracking analysis (NTA, NanoSight NTA v3.2) after 1:1000 dilution with particle-free PBS [68], and the characteristic markers (CD9, TSG101, and HSP70) of hBMSC-EVs were verified using Western blotting. The effect of hBMSC-EVs on the proliferation and migration behavior of the OS cell line MG63 (ATCC, VA, USA) was determined using an MTT assay and a scratch test. The effects of three different concentrations of hBMSC-EVs (3 μg/mL, 30 μg/mL, and 300 μg/mL) on the proliferation of the OS cell line MG63 were determined using MTT assays on day 2 and day 4. The effects of different concentrations of hBMSC-EVs (3 μg/mL, 30 μg/mL, and 300 μg/mL) on the migration of MG63 cells were determined using a scratch test in serum-free medium at 72 h.
5.1.2. Extraction and identification of dOsEM
Six fresh human OS tissue samples (preserved in medium) were first minced with ultrasonic mechanical disruption and placed in 0.25 % trypsin for 6 h while stirring at 250 rpm, washed three times with water, treated with 100 mL of 70 % ethanol at 250 rpm and stirred for 10 h, and washed with 3 % H2O2 for 15 min. After washing three times, 1 % Triton X-100 and 0.26 % EDTA/Tris were added for 6 h. Next, the tissue was washed three times with water for 15 min, treated with 0.1 % peracetic acid and 4 % ethanol for 2 h, washed with double-distilled water for 15 min three times, and then freeze-dried. The DNA content in the acellular matrix was quantified using a NanoDrop (Thermo Fisher, MA, USA), and the aforementioned components were quantified using a Human GAGs ELISA Kit (FineTest, Wuhan, China) and Collagen ELISA Quantification Kit (Abcam, Cambridge, UK). Furthermore, qualitative and quantitative analyses of the matrix components were performed using the protein spectrum. The XPS spectrum was measured on K-Alpha (Thermal Scientific, USA).
5.1.3. Biomass spectrometry and protein profiling for dOsEM and hBMSC-EVs
Biomass spectrometry was used to analyze the protein macromolecules in dOsEM and hBMSC-EVs. After cleavage the protein from three separated samples, the proteins were preliminarily separated by SDS‒PAGE electrophoresis, and a sequencing-grade trypsin solution was added to react overnight at 37 °C. Data were then quantitively analyzed using MaxQuant software.
5.1.4. Preparation of the micro-OS
Lyophilized dOsEM and pepsin (Sigma, St. Louis, MO, USA) were dissolved at a ratio of 10:1 with stirring for 48 h. The pH was adjusted to 7.4 using NaOH, and a final concentration of 6 mg/mL was reached. According to our previous study [69], the bioink for micro-OS was produced using the components and proportions listed in Table 1. Furthermore, the rheological and mechanical properties of the bioink are described in Fig. 1G and H.
Table 1.
Composition of printable dOsEM-fibrin hydrogels.
| EV concentration | dOsEM | Fibrinogen | Gelatin | Glycerol | MG63 cell density | |
|---|---|---|---|---|---|---|
| hBMSC-EV-dOsEM-fibrin | 300 μg/mL | 6 mg/mL | 20 mg/mL | 20 mg/mL | 10 % | 1.0 × 107/mL |
| dOsEM-fibrin | None | 6 mg/mL | 20 mg/mL | 20 mg/mL | 10 % | 1.0 × 107/mL |
Briefly, the rheological behavior of the dOsEM-fibrin solution (3%–5%) with different concentrations of gelatin (1%–3%) (Sigma, St. Louis, MO, USA) was characterized using an HR-2 Discovery rheometer (TA Instruments, Newcastle, DE, USA) with a 25-mm steel parallel plate geometry [44]. The gap between the plate and the surface of the dOsEM-fibrin solution was 105 μm. Strain was measured from 0.01 to 100 at an angular frequency of 6.28 rad/s. The rheometer was operated using the programmable software of the instrument. Data were collected from triplicate samples, and the temperature of the solution was maintained at 20 °C. The MG63 cell proliferation activity in dOsEM-fibrin was compared with that in four other typical hydrogels, GelMA hydrogel, hyaluronan/Gelin/PEGDA hydrogel, collagen hydrogel, and fibrin hydrogel, using CCK-8 assays on days 1, 3, and 5. A 3D printer (Regenove Bio-Printer-WS, China) was used to construct 3D-printed hBMSC-EV-loaded micro-OSs by extruding and stacking the hydrogel layer-by-layer. After installing the cartridge to the head of the bioprinter, the printing speed was set to 3 mm/s, and the printing figure was set to be a cuboid (10 × 10 × 5 mm). In addition, based on the results of previous studies of oxygen diffusion, 1.0-mm printing spacing models were used. Then, the micro-OS was formed using extruded biomaterials driven by air pressure. At least three replicate samples of each hydrogel scaffold were used. After printing, the 3D-printed micro-OS samples were added to a thrombin (200 IU/mL, Sigma, St. Louis, MO, USA) solution at 37 °C for crosslinking for 30 min. The Young's Modulus of the micro-OS was performed on an Instron 5982 universal testing system.
5.2. Characterization of physicochemical properties of the micro-OS
5.2.1. Cell viability and proliferation analyses
The proliferation of the OS cell line MG63 (ATCC, VA, USA) in the hydrogel was measured using a CCK-8 Lit (Biosharp, China) and a Live/Dead Viability Kit (Invitrogen, USA). The CCK-8 reagent was diluted in the medium at a ratio of 1:10. The medium inside the well plate was aspirated. Then, 300 μL of the CCK-8 diluted solution was added and incubated at room temperature for 2 h. At 1, 3, 5, and 7 days, the absorbance was assessed at 450 nm inside the well plate using four parallel samples in each group. For live/dead staining, the samples were treated with 5 μL/mL calcein AM and 2 μL/mL propidium iodide (PI) for 45 min at 37 °C. The fluorescence was examined using a fluorescence microscope (X-Cite® 120 LED Boost, Olympus). Six random fields for six samples were used to analyze viability using ImageJ software.
5.2.2. Ultrastructure analysis
After culturing the scaffolds for 24 h, the medium was removed, and the scaffolds were washed three times with PBS. Next, 2.5 % paraformaldehyde was added for fixation at room temperature for 60 min. The hydrogel scaffolds were then washed three times with PBS and afterward with ddH2O. An ethanol gradient was used for gradient dehydration treatment, and the samples were dried. Finally, the samples were sputter coated with gold, and the surface morphology of the scaffold was observed using scanning electron microscopy (SEM, FlexSEM 1000; Hitachi, Tokyo, Japan). The pseudopodium extensions for cell adhesion on the scaffold were also observed through SEM.
5.2.3. RNA-seq and RT‒PCR
RNA-seq technology was used to analyze the key regulatory genes in MSC-EVs and their downstream activated signaling pathways. RNA was obtained from the 2D culture group, 3D micro-OS group, and xenograft group to obtain RNA using the TRIzol cleavage method. A TruSeq Strand mRNA Sample Preparation Kit was used to construct the cDNA library. The cDNA in the library was subsequently sequenced using an Illumina HiSeq X Ten detection platform. Each group of three replicate sample reads was recorded. The raw data were processed using Trimmomatic software and analyzed and compared using the DESeq (2012) R package to obtain the differential gene expression results, including biological processes (BP), cell components (CC), molecular functions (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. For RT‒PCR, total RNA was isolated using an RNeasy Mini Kit (QIAGEN, Germantown, PA, USA). qPCR analysis of the genes was performed with the probes listed in Supplementary Table S1. All qPCR experiments were performed on a QuantStudio 3 PCR system (Thermo Scientific, Waltham, MA, USA).
5.2.4. Histology and immunofluorescence
The sample was washed three times with PBS, and 2.5 % paraformaldehyde was added. The sample was fixed at room temperature for 60 min. Then, the sample was washed three times with PBS containing 0.1 % Triton X-100 and blocked using BSA (Dako, Santa Clara, CA, USA) at room temperature. Next, the sample was incubated with the primary antibodies (Supplementary Table S2) overnight at 4 °C and then incubated with the fluorescence-conjugated secondary antibody at room temperature. DAPI (Invitrogen) was used to stain the nuclei. A confocal laser scanning microscope (X-Cite® 120 LED Boost, Olympus) was used to image the cells.
5.3. Preparation and characterization of the OOC model
5.3.1. Isolation of patient OS cells
The primary OS specimens were freshly isolated from the OS patient (i), and after being minced with scissors (ii), OS tissues were digested with Tissue Dissociation Solution (Cytowise, Shanghai, China) under magnetic stirring (iii). The digested cells were then separated from the digestion solution by centrifugation at 1300 rpm for 10 min (iv). The OS specimens were also used as positive controls for the OOC and xenograft models.
5.3.2. Preparation of the OOC model using the 3D-printed micro-OS
The hBMSC-EV-loaded dOsEM-fibrin scaffolds loaded with MG63 cell line or patient-derived OS cells were then constructed using 3D bioprinting. The glass slides, adhesive films (DSF), and PMMA were stacked and firmly fixed together after printing. The top assembly consisted of plasma-bound adhesive and a PDMS cover. The port on the lid was fitted using polytetrafluoroethylene tubing (Cole Parmer, Vernon Hills, IL), and the chip was connected to a micro peristaltic pump (MP2 Precision, Elemental Scientific, Inc., Omaha, NE) circuit. Then, the pipe from the inlet port was connected to the medium container using the PVC pipe, and the outlet pipe was set to discharge into the container, thus establishing a closed loop system. PVC tubing was installed on the micro peristaltic pump to initiate medium recirculation in the system. The 3D-printed OS tissue was grown in a simulated circulatory system by filling each medium reservoir with 1.6 mL of medium and then pumping at a very low flow rate of 4 μL/min for 48 h, based on our previous research [9,10].
5.3.3. Characterization of the OOC model
The effectiveness of the culture system was verified based on OS cell proliferation, migration, and apoptosis in the chip through CaAM/PI staining, MTT assay and IHC staining (Supplementary Table S2). 2D: MG63 cells under 2D culture for 48 h; Static: MG63 cells under the OOC culture system without perfusion for 48 h; OOC: MG63 cells under the OOC culture system with perfusion for 48 h; Xenograft: MG63 cell mouse xenograft models 2 weeks after seeding. Patient: Fresh OS tissues from patient.
5.4. Construction of the CXCR4–CXCL12 complex by protein‒protein docking
The crystal structures of CXCR4 and CXCL12 were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/): PDB ID 3ODU for CXCR4 and 2J7Z for CXCL12. The proteins were then prepared for docking by removing water molecules and ligands. Hydrogen was added before performing protein‒protein docking.
First, rigid docking was carried out through the ClusPro online server (https://cluspro.bu.edu/) to adjust the initial conformations of the two proteins. Then, based on the results of rigid docking, RosettaDock was used for flexible docking. The results were selected by screening with Rosetta's built-in score module. Then, we used LigPlot to analyze intermolecular interactions. The binding energy between CXCL12 and CXCR4 was calculated by the Interface analyzer module from Rosetta software. Finally, the CXCR4–CXCL12 complex was visualized in PyMOL molecular graphics software to identify the interactive residues.
5.5. Molecular docking of the CXCR4–CXCL12 complex
We then used the optimized the CXCR4–CXCL12 complex obtained by protein‒protein docking as the protein structure for molecular docking. The 3D structure of plerixafor was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/).
First, AutodockTool was utilized to hydrogenate the predicted protein structure and assign atom types. Then, we performed molecular docking with Autodock4 based on the protein structure output from AutodockTool. Based on the results of the key residues for the interaction between CXCR4 and CXCL12, a local docking mode was adopted. The Autodock4 program automatically predicted the binding free energy according to an algorithm based on the AMBER force field. The final confirmation of the ligand was chosen according to the predicted binding free energy. The interaction force between CXCL12 and CXCR4 was analyzed by Ligplot software, and the 3D ligand‒receptor conformation was displayed by PyMOL software.
5.6. Preparation of a tumor-bearing nude mouse model and drug evaluation
MG63 cells were seeded subcutaneously in BALB/c nude mice (JSJ Laboratory, Shanghai, China) at a cell density of 5 × 106/50 μL. After two weeks of tumor formation, the body weight and tumor size of the nude mice were recorded, and different doses of drugs were administered. Six mice were included in each group. The drugs were administered intraperitoneally (DOX: 3 mg/kg [70,71], cisplatin: 6 mg/kg [72], AMD3100: 5 mg/kg mouse weight [73,74]) (Fig. 7A), and the body weight and tumor size of the nude mice were recorded every two days.
The DOX concentration inside the OS chip and the tumor-bearing mouse model were determined at different standard concentrations using a microplate spectrophotometer (Multiskan GO, Thermo Scientific, USA). The excitation and emission wavelengths were 470 and 590 nm, respectively. Linear regression was performed for the absorbance values and the DOX concentration to obtain the absorbance–concentration standard curve y = 0.4661x – 14.313 (R2 = 0.9992). Then, the measured absorbance of each sample solution was converted to the concentration according to the standard curves. The pharmacokinetic and pharmacodynamic properties of DOX inside the OS tissues were determined using Phoenix 64 (version 8.1.0.3530) based on the DOX concentration in the blood and tumor.
HL_Lambda_z: Terminal half-life = ln (2)/Lambda Z;
Cmax: Maximum observed concentration;
AUClast: Area under the curve from the time of dosing to the time of the last measurable (positive) concentration (Tlast);
AUCINF_obs: AUC from time of dosing extrapolated to infinity, based on the last observed concentration (_obs) or last predicted concentration;
Vz_F_obs: Volume of distribution based on the terminal phase;
Cl_F_obs: Total body clearance for extravascular administration;
MRTINF_obs: Mean residence time (MRT) extrapolated to infinity.
5.7. Computational simulation of DOX release
We used the model developed in this study to determine the related parameters in the OOC system. The transport of DOX in the chips was simulated using the finite element software COMSOL Multiphysics (version 5.4). The permeability of DOX in an OOC system with cells was expected to be lower than the permeability in medium, as described by the obstruction theory, because dOsEM is composed of biofibers that can hinder DOX transport, and the DOX diffusivity of cells suspended in the gel is less than that of cells in medium (scaffold porosity). Therefore, the diffusion coefficients in the hydrogel and water were assumed to be 2.3 × 10−10 m2 s− 1 and 3.8 × 10−9 m2 s− 1, respectively, based on our previous studies [10,50]. Then, simulations were performed using the concentration data measured at steady-state points to determine the release rate of DOX and to yield the best fit between the measured and predicted DOX concentrations inside the hydrogel [10].
5.8. Statistical analyses
GraphPad Prism and SPSS software were used to perform statistical analyses on functional and histological data. The data are displayed as the mean ± standard deviation. To compare two groups, significant differences between values were calculated using Student's t tests. To compare more than two groups, one-way analysis of variance (ANOVA) was used. When the ANOVA results revealed a significant difference, a Newman‒Keuls test was performed. Significance thresholds are indicated by *p < 0.05, **p < 0.01, and ***p < 0.001.
Ethics approval
Animals: The experiments were approved by the Ethics Committee of Shanghai Ninth People's Hospital, Shanghai, China (SH9H-2021-A967-SB). Human: The experiments were approved by the Ethics Committee of Shanghai Ninth People's Hospital, Shanghai, China (SH9H-2023-TK44-1).
Funding
The authors wish to acknowledge the Shanghai Pujiang Program (21PJ1409200); the China Postdoctoral Science Foundation (2022M722122); Three-year Action Plan of Shenkang Development Center (SHDC2020CR2019B); Biomaterials and Regenerative Medicine Institute Cooperative Research Project, Shanghai Jiao Tong University School of Medicine (2022LHB07, 2022LHA01); Shanghai Frontiers Science Center of Degeneration and Regeneration in Skeletal System; the Fundamental Research Funds for the Central Universities (YG2023LC07); Clinical Research Program of Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine (JYLJ202122); the Project of Biobank from Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (YBKB202116) and the National Scientific Foundation of China (82171993, 81972058, 82130073).
Data and materials availability
The datasets generated for this study are available on request from either of the corresponding authors.
CRediT authorship contribution statement
Conceptualization: ZL and KD. Methodology: ZL, XM, CZ, BS, AS, AA, JG, SA. Investigation: ZL, XW, JG. Visualization: ZL, CZ, JG. Funding acquisition: ZL and KD. Project administration: ZL. Supervision: AS, AA, YH, JZ. Writing ? original draft: ZL. Writing ? review & editing: ZL, CZ, AS.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to Shiny Amala Priya Rajan, Yanan Sun and Mi Chen for useful discussions and technical assistance.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.12.005.
Contributor Information
Yongqiang Hao, Email: hyq_9hospital@hotmail.com.
Jie Zhao, Email: profzhaojie@126.com.
Kerong Dai, Email: krdai@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Gengenbacher N., Singhal M., Augustin H.G. Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat. Rev. Cancer. 2017;17(12):751–765. doi: 10.1038/nrc.2017.92. [DOI] [PubMed] [Google Scholar]
- 2.Padrón J.M., van der Wilt C.L., Smid K., Smitskamp-Wilms E., Backus H.H., Pizao P.E., Giaccone G., Peters G.J. The multilayered postconfluent cell culture as a model for drug screening. Crit. Rev. Oncol.-Hematol. 2000;36(2–3):141–157. doi: 10.1016/s1040-8428(00)00083-4. [DOI] [PubMed] [Google Scholar]
- 3.Feng D., Li Z., Yang L., Liang H., He H., Liu L., Zhang W. BMSC-EV-derived lncRNA NORAD facilitates migration, invasion, and angiogenesis in osteosarcoma cells by regulating CREBBP via delivery of miR-877-3p. Oxid. Med. Cell. Longev. 2022:2022. doi: 10.1155/2022/8825784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du G.Y., He S.W., Zhang L., Sun C.X., Mi L.D., Sun Z.G. Hesperidin exhibits in vitro and in vivo antitumor effects in human osteosarcoma MG-63 cells and xenograft mice models via inhibition of cell migration and invasion, cell cycle arrest and induction of mitochondrial-mediated apoptosis. Oncol. Lett. 2018;16(5):6299–6306. doi: 10.3892/ol.2018.9439. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Gould S.E., Junttila M.R., de Sauvage F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015;21(5):431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 6.Killion J.J., Radinsky R., Fidler I.J. Orthotopic models are necessary to predict therapy of transplantable tumors in mice. Cancer Metast Rev. 1998;17:279–284. doi: 10.1023/a:1006140513233. [DOI] [PubMed] [Google Scholar]
- 7.Vargo-Gogola T., Rosen J.M. Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer. 2007;7(9):659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 8.Rajan S.A.P., Aleman J., Wan M., Zarandi N.P., Nzou G., Murphy S., Bishop C.E., Sadri-Ardekani H., Shupe T., Atala A. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020;106:124–135. doi: 10.1016/j.actbio.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleman J., George S.K., Herberg S., Devarasetty M., Porada C.D., Skardal A., Almeida Porada G. Deconstructed microfluidic bone marrow on‐A‐chip to study normal and malignant hemopoietic cell–niche interactions. Small. 2019;15(43) doi: 10.1002/smll.201902971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z., Rajan S.A.P., Song Q., Zhao Y., Wan M., Aleman J., Skardal A., Bishop C., Atala A., Lu B. 3D scaffold-free microlivers with drug metabolic function generated by lineage-reprogrammed hepatocytes from human fibroblasts. Biomaterials. 2021;269 doi: 10.1016/j.biomaterials.2021.120668. [DOI] [PubMed] [Google Scholar]
- 11.McAleer C.W., Pointon A., Long C.J., Brighton R.L., Wilkin B.D., Bridges L.R., Narasimhan Sriram N., Fabre K., McDougall R., Muse V.P. On the potential of in vitro organ-chip models to define temporal pharmacokinetic-pharmacodynamic relationships. Sci Rep-UK. 2019;9(1):1–14. doi: 10.1038/s41598-019-45656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marturano-Kruik A., Nava M.M., Yeager K., Chramiec A., Hao L., Robinson S., Guo E., Raimondi M.T., Vunjak-Novakovic G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA. 2018;115(6):1256–1261. doi: 10.1073/pnas.1714282115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19(2):65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 14.Mitxelena-Iribarren O., Zabalo J., Arana S., Mujika M. Improved microfluidic platform for simultaneous multiple drug screening towards personalized treatment. Biosens. Bioelectron. 2019;123:237–243. doi: 10.1016/j.bios.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y., Bao Q., Yu S., Liu Q., Xie Y., Li X., Liu Y., Shen Y. A novel microfluidic chip for fast, sensitive quantification of plasma extracellular vesicles as biomarkers in patients with osteosarcoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.709255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteiro C.F., Custódio C.A., Mano J.F. Three‐dimensional osteosarcoma models for advancing drug Discovery and development. Advanced Therapeutics. 2019;2(3) [Google Scholar]
- 17.Becquart P., Cambon-Binder A., Monfoulet L., Bourguignon M., Vandamme K., Bensidhoum M., Petite H., Logeart-Avramoglou D. Ischemia is the prime but not the only cause of human multipotent stromal cell death in tissue-engineered constructs in vivo. Tissue Eng. 2012;18(19–20):2084–2094. doi: 10.1089/ten.TEA.2011.0690. [DOI] [PubMed] [Google Scholar]
- 18.Heinrich M.A., Liu W., Jimenez A., Yang J., Akpek A., Liu X., Pi Q., Mu X., Hu N., Schiffelers R.M. 3D bioprinting: from benches to translational applications. Small. 2019;15(23) doi: 10.1002/smll.201805510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garimella R., Washington L., Isaacson J., Vallejo J., Spence M., Tawfik O., Rowe P., Brotto M., Perez R. Extracellular membrane vesicles derived from 143B osteosarcoma cells contain pro-osteoclastogenic cargo: a novel communication mechanism in osteosarcoma bone microenvironment. Transl Oncol. 2014;7(3):331–340. doi: 10.1016/j.tranon.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu P., Takai K., Weaver V.M., Werb Z. Extracellular matrix degradation and remodeling in development and disease. CSH Perespect Biol. 2011;3(12):a5058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMillin D.W., Negri J.M., Mitsiades C.S. The role of tumour–stromal interactions in modifying drug response: challenges and opportunities. Nat. Rev. Drug Discov. 2013;12(3):217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- 23.Maman S., Witz I.P. A history of exploring cancer in context. Nat. Rev. Cancer. 2018;18(6):359–376. doi: 10.1038/s41568-018-0006-7. [DOI] [PubMed] [Google Scholar]
- 24.Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 25.De Palma M., Biziato D., Petrova T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer. 2017;17(8):457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 26.Valkenburg K.C., De Groot A.E., Pienta K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018;15(6):366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun B., Han Y., Jiang W., Dai K. 3D printing bioink preparation and application in cartilage tissue reconstruction in vitro. J. Shanghai Jiaot. Univ. 2021;26(3):267–271. [Google Scholar]
- 28.Roorda B.D., ter Elst A., Kamps W.A., de Bont E.S. Bone marrow-derived cells and tumor growth: contribution of bone marrow-derived cells to tumor micro-environments with special focus on mesenchymal stem cells. Crit. Rev. Oncol.-Hematol. 2009;69(3):187–198. doi: 10.1016/j.critrevonc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Shinagawa K., Kitadai Y., Tanaka M., Sumida T., Kodama M., Higashi Y., Tanaka S., Yasui W., Chayama K. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int. J. Cancer. 2010;127(10):2323–2333. doi: 10.1002/ijc.25440. [DOI] [PubMed] [Google Scholar]
- 30.Fatima F., Nawaz M. Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Non-coding RNA. 2017;3(1):10. doi: 10.3390/ncrna3010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W., Huang L., Li Y., Zhang X., Gu J., Yan Y., Xu X., Wang M., Qian H., Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315(1):28–37. doi: 10.1016/j.canlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Vrijsen K.R., Sluijter J., Schuchardt M., Van Balkom B., Noort W.A., Chamuleau S., Doevendans P. Cardiomyocyte progenitor cell‐derived exosomes stimulate migration of endothelial cells. J. Cell Mol. Med. 2010;14(5):1064–1070. doi: 10.1111/j.1582-4934.2010.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xin H., Li Y., Liu Z., Wang X., Shang X., Cui Y., Zhang Z.G., Chopp M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cell. 2013;31(12):2737–2746. doi: 10.1002/stem.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T., Lee Y.W., Rui Y.F., Cheng T.Y., Jiang X.H., Li G. Bone marrow-derived mesenchymal stem cells promote growth and angiogenesis of breast and prostate tumors. Stem Cell Res. Ther. 2013;4(3):1–15. doi: 10.1186/scrt221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He H., Ding M., Li T., Zhao W., Zhang L., Yin P., Zhang W. Bone mesenchymal stem cell-derived extracellular vesicles containing NORAD promote osteosarcoma by miR-30c-5p. Lab. Invest. 2022;102(8):826–837. doi: 10.1038/s41374-021-00691-6. [DOI] [PubMed] [Google Scholar]
- 36.Tchoukalova Y.D., Hintze J.M., Hayden R.E., Lott D.G. Tracheal decellularization using a combination of chemical, physical and bioreactor methods. Int. J. Artif. Organs. 2018;41(2):100–107. doi: 10.5301/ijao.5000648. [DOI] [PubMed] [Google Scholar]
- 37.Xie M., Shi Y., Zhang C., Ge M., Zhang J., Chen Z., Fu J., Xie Z., He Y. In situ 3D bioprinting with bioconcrete bioink. Nat. Commun. 2022;13(1):1–12. doi: 10.1038/s41467-022-30997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maloney E., Clark C., Sivakumar H., Yoo K., Aleman J., Rajan S.A., Forsythe S., Mazzocchi A., Laxton A.W., Tatter S.B. Immersion bioprinting of tumor organoids in multi-well plates for increasing chemotherapy screening throughput. Micromachines-Basel. 2020;11(2):208. doi: 10.3390/mi11020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.H., Kim I., Seol Y., Ko I.K., Yoo J.J., Atala A., Lee S.J. Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function. Nat. Commun. 2020;11(1):1025. doi: 10.1038/s41467-020-14930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y., Zhou Y., Zhang R., Wen L., Wu K., Li Y., Yao Y., Duan R., Jia Y. Bone mesenchymal stem cell-derived extracellular vesicles promote recovery following spinal cord injury via improvement of the integrity of the blood-spinal cord barrier. Front Neurosci-Switz. 2019;13:209. doi: 10.3389/fnins.2019.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominkuš P.P., Stenovec M., Sitar S., Lasič E., Zorec R., Plemenitaš A., Žagar E., Kreft M., Lenassi M. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim. Biophys. Acta Biomembr. 2018;1860(6):1350–1361. doi: 10.1016/j.bbamem.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Yao X., Lyu P., Yoo K., Yadav M.K., Singh R., Atala A., Lu B. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J. Extracell. Vesicles. 2021;10(5) doi: 10.1002/jev2.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu S., Xing H., Zhang J., Zhu Z., Yin Y., Zhang N., Qi Y. Mesenchymal stem cell-derived extracellular vesicles: immunomodulatory effects and potential applications in intervertebral disc degeneration. Stem Cells Int. 2022:2022. doi: 10.1155/2022/7538025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L., Varkey M., Jorgensen A., Ju J., Jin Q., Park J.H., Fu Y., Zhang G., Ke D., Zhao W. Bioprinting small diameter blood vessel constructs with an endothelial and smooth muscle cell bilayer in a single step. Biofabrication. 2020;12(4) doi: 10.1088/1758-5090/aba2b6. [DOI] [PubMed] [Google Scholar]
- 45.Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Chun R., Kurzman I.D., Couto C.G., Klausner J., Henry C., MacEwen E.G. Cisplatin and doxorubicin combination chemotherapy for the treatment of canine osteosarcoma: a pilot study. J. Vet. Intern. Med. 2000;14(5):495–498. doi: 10.1892/0891-6640(2000)014<0495:cadccf>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 47.Kudawara I., Aoki Y., Ueda T., Araki N., Naka N., Nakanishi H., Matsumine A., Ieguchi M., Mori S., Myoui A. Neoadjuvant and adjuvant chemotherapy with high-dose ifosfamide, doxorubicin, cisplatin and high-dose methotrexate in non-metastatic osteosarcoma of the extremities: a phase II trial in Japan. J Chemothraphy. 2013;25(1):41–48. doi: 10.1179/1973947812Y.0000000055. [DOI] [PubMed] [Google Scholar]
- 48.González Díaz E.C., Lee A.G., Sayles L.C., Feria C., Sweet Cordero E.A., Yang F. A 3D osteosarcoma model with bone‐mimicking cues reveals a critical role of bone mineral and informs drug Discovery. Adv Healthc Mater. 2022;11(17) doi: 10.1002/adhm.202200768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha S., Ayushman M., Tong X., Yang F. Dynamically crosslinked poly (ethylene‐glycol) hydrogels reveal a critical role of viscoelasticity in modulating glioblastoma fates and drug responses in 3D. Adv Healthc Mater. 2023;12(1) doi: 10.1002/adhm.202202147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Z., Jiang X., Chen M., Feng L., Kang Y.J. An oxygen-releasing device to improve the survival of mesenchymal stem cells in tissue engineering. Biofabrication. 2019;11(4) doi: 10.1088/1758-5090/ab332a. [DOI] [PubMed] [Google Scholar]
- 51.Nunes A.S., Barros A.S., Costa E.C., Moreira A.F., Correia I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019;116(1):206–226. doi: 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- 52.Barbosa M.A., Xavier C.P., Pereira R.F., Petrikaitė V., Vasconcelos M.H. 3D cell culture models as recapitulators of the tumor microenvironment for the screening of anti-cancer drugs. Cancers. 2022;14(1):190. doi: 10.3390/cancers14010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T., Li W., Zhang Y., Xu X., Qiang L., Miao W., Yue X., Jiao X., Zhou X., Ma Z. Bioprinted constructs that simulate nerve–bone crosstalk to improve microenvironment for bone repair. Bioact. Mater. 2023;27:377–393. doi: 10.1016/j.bioactmat.2023.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pablos J.L., Amara A., Bouloc A., Santiago B., Caruz A., Galindo M., Delaunay T., Virelizier J.L., Arenzana-Seisdedos F. Stromal-cell derived factor is expressed by dendritic cells and endothelium in human skin. Am. J. Pathol. 1999;155(5):1577–1586. doi: 10.1016/S0002-9440(10)65474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muehlberg F.L., Song Y., Krohn A., Pinilla S.P., Droll L.H., Leng X., Seidensticker M., Ricke J., Altman A.M., Devarajan E. Tissue-resident stem cells promote breast cancer growth and metastasis. Carcinogenesis. 2009;30(4):589–597. doi: 10.1093/carcin/bgp036. [DOI] [PubMed] [Google Scholar]
- 56.Halpern J.L., Kilbarger A., Lynch C.C. Mesenchymal stem cells promote mammary cancer cell migration in vitro via the CXCR2 receptor. Cancer Lett. 2011;308(1):91–99. doi: 10.1016/j.canlet.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Jiang H., Guo L., Wang S., Cheng W., Wan L., Zhang Z., Xing L., Zhou Q., Yang X. SDF-1 secreted by mesenchymal stem cells promotes the migration of endothelial progenitor cells via CXCR4/PI3K/AKT pathway. J. Mol. Histol. 2021;52(6):1155–1164. doi: 10.1007/s10735-021-10008-y. [DOI] [PubMed] [Google Scholar]
- 58.Furusato B., Rhim J.S. CXCR4 and cancer. Chemokine Receptors in Cancer. 2009:31–45. [Google Scholar]
- 59.Shi Y., Fang W. Hypoxia-inducible factor-1 in tumour angiogenesis. World J. Gastroenterol. 2004;10(8):1082. doi: 10.3748/wjg.v10.i8.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang X., Wang C., Fitch S., Yang F. Targeting tumor hypoxia using nanoparticle-engineered CXCR4-overexpressing adipose-derived stem cells. Thearnostics. 2018;8(5):1350. doi: 10.7150/thno.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lortat-Jacob H. The molecular basis and functional implications of chemokine interactions with heparan sulphate. Curr. Opin. Struct. Biol. 2009;19(5):543–548. doi: 10.1016/j.sbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Purcell B.P., Elser J.A., Mu A., Margulies K.B., Burdick J.A. Synergistic effects of SDF-1α chemokine and hyaluronic acid release from degradable hydrogels on directing bone marrow derived cell homing to the myocardium. Biomaterials. 2012;33(31):7849–7857. doi: 10.1016/j.biomaterials.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., Tang R. Localized delivery of chemokine for in vitro manipulation of hepatocellular carcinoma cell behaviors during the epithelial–mesenchymal transition. J. Biomater. Appl. 2018;32(7):945–956. doi: 10.1177/0885328217745774. [DOI] [PubMed] [Google Scholar]
- 64.Wagstaff A.J. Plerixafor. DRUGS. 2009;69(3):319–326. doi: 10.2165/00003495-200969030-00007. [DOI] [PubMed] [Google Scholar]
- 65.Dong Y., Liang G., Yuan B.O., Yang C., Gao R., Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor Biol. 2015;36(3):1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 66.Zhang M., Qiu L., Zhang Y., Xu D., Zheng J.C., Jiang L. CXCL12 enhances angiogenesis through CXCR7 activation in human umbilical vein endothelial cells. Sci Rep-UK. 2017;7(1):1–9. doi: 10.1038/s41598-017-08840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fallica B., Maffei J.S., Villa S., Makin G., Zaman M. 2012. Alteration of Cellular Behavior and Response to PI3K Pathway Inhibition by Culture in 3D Collagen Gels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou M., Weber S.R., Zhao Y., Chen H., Sundstrom J.M. Methods for exosome isolation and characterization. Exosomes. 2020:23–38. [Google Scholar]
- 69.Kim J.H., Kim I., Seol Y., Ko I.K., Yoo J.J., Atala A., Lee S.J. Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-14930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma H., He C., Cheng Y., Yang Z., Zang J., Liu J., Chen X. Localized co-delivery of doxorubicin, cisplatin, and methotrexate by thermosensitive hydrogels for enhanced osteosarcoma treatment. ACS Appl Mater Inter. 2015;7(49):27040–27048. doi: 10.1021/acsami.5b09112. [DOI] [PubMed] [Google Scholar]
- 71.Higuchi T., Sugisawa N., Miyake K., Oshiro H., Yamamoto N., Hayashi K., Kimura H., Miwa S., Igarashi K., Bouvet M. The combination of olaratumab with doxorubicin and cisplatinum regresses a chemotherapy-resistant osteosarcoma in a patient-derived orthotopic xenograft mouse model. Transl Oncol. 2019;12(9):1257–1263. doi: 10.1016/j.tranon.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu N.F., Yamamoto J., Aoki Y., Masaki N., Samonte C., Wu J., Bouvet M., Hoffman R.M. The combination of cisplatinum and doxorubicin regressed primary osteosarcoma of the breast in a PDOX mouse model. Anticancer Res. 2021;41(10):4715–4718. doi: 10.21873/anticanres.15285. [DOI] [PubMed] [Google Scholar]
- 73.Omer-Javed A., Pedrazzani G., Albano L., Ghaus S., Latroche C., Manzi M., Ferrari S., Fiumara M., Jacob A., Vavassori V. Mobilization-based chemotherapy-free engraftment of gene-edited human hematopoietic stem cells. Cell. 2022;185(13):2248–2264. doi: 10.1016/j.cell.2022.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chien S., Zhao X., Papayannopoulou T., Appelbaum F.R., Becker P.S. The CXCR4 inhibitor plerixafor effectively mobilizes primary AML in NODscid IL2Rγc−/− xenografts and markedly reduces but does not eradicate leukemia in combination with Cytarabine+/− clofarabine chemotherapy. Blood. 2011;118(21):1432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.