Abstract
Recent studies have revealed the impact of microRNAs (miRNAs) in the carcinogenic process. miR-424 is a miRNA whose role in this process is being to be identified. Experiments in the ovarian cancer, cervical cancer, hepatocellular carcinoma, neuroblastoma, breast cancer, osteosarcoma, intrahepatic cholangiocarcinoma, prostate cancer, endometrial cancer, non-small cell lung cancer, hemangioma and gastric cancer have reported down-regulation of miR-424. On the other hand, this miRNA has been found to be up-regulated in melanoma, laryngeal and esophageal squamous cell carcinomas, glioma, multiple myeloma and thyroid cancer. Expression of this miRNA is regulated by methylation status of its promoter. Besides, LINC00641, CCAT2, PVT1, LIN00657, LINC00511 and NNT-AS1 are among lncRNAs that act as molecular sponges for miR-424, thus regulating its expression. Moreover, several members of SNHG family of lncRNAs have been found to regulate expression of miR-424. This miRNA is also involved in the regulation of E2F transcription factors. The current review aims at summarization of the role of miR-424 in the process of cancer evolution and its impact on clinical outcome of patients in order to find appropriate markers for malignancies.
Keywords: miR-424, Cancer, Biomarker
Introduction
MicroRNAs (miRNAs) are transcripts with sizes around 23 nt that can influence expression of genes at post-transcriptional level. These small, highly conserved transcripts are transcribed by RNA polymerases II and III. The miRNA precursors generated by these polymerases go through a group of cleavage actions to make mature miRNAs [1]. miRNA precursors are lengthy polycistronic RNAs that share some features with mRNAs since they possess distinctive 5′ and 3′ borders, 7-methyl guanylate caps and poly(A) tails.
The function of miRNAs in the regulation of genes expression is accomplished via the RNA-induced silencing complex (RISC) [2]. After assemblage into RISC, miRNAs activate this complex to target mRNAs in a specific manner [1]. miRNAs which control oncogenes and tumor suppressor genes, are differentially expressed in various human malignancies and play a central role in all cancer hallmarks, especially in their real targets [3–6]. Moreover, miRNAs differ in their transcriptional units and the mechanisms of their regulation within genomic loci. Those being located within an intron of a host gene are transcribed in the identical orientation with primary transcript by the promoter of the host gene [7]. On the other hand, miRNAs located in the intergenic loci have their own promoters [7, 8].
miR-424 is encoded by a gene located on chr Xq26.3. This miRNA has been demonstrated to be dysregulated in different cancers. Notably, dysregulation of miR-424 in cancer samples have been associated with invasive behavior of malignant cells. However, different studies have reported various results regarding its expression in different cancers. Mechanistically, several lncRNAs act as molecular sponge for miR-424 to regulate its expression. The current review aims at summarization of the role of miR-424 in the process of cancer evolution and its impact on clinical outcome of patients in order to find appropriate markers for malignancies.
Cancer cell lines
An experiment in neuroblastoma cell lines has revealed down-regulation of miR-424 and up-regulation of its target gene DCLK1 in these cells compared with normal spongiocyte cells. Mechanistical studies has confirmed that the role of miR-424 in suppression of cell viability, invasive properties, and epithelial-mesenchymal transition (EMT) is mediated through targeting DCLK1. In fact, DCLK1 could partially reverse function of miR-424 in neuroblastoma cells [9].
In MG-63 and SaOS2 osteosarcoma cells, expression of miR-424-5p has been increased upon treatment with melatonin leading to inhibition of VEGFA. Moreover, melatonin could suppress neoangiogenesis, affecting proliferation and migration of neighboring endothelial cells as well as release of angiogenic growth factors. It has also induced morphological changes in blood vessels, and. Taken together, melatonin has a tumor suppressive role through influencing miR-424-5p/VEGFA axis [10].
In silico studies in glioblastoma have predicted a tumor suppressor role for miR-424. This miRNA has also been predicted to target several genes from the ERBB signaling pathway that are activated in the majority of glioblastoma patients. Cell line studies have also confirmed the impact of miR-424 overexpression in suppression of proliferation and migratory potential of glioblastoma cells. Moreover, miR-424 has been shown to promote apoptosis and induce cell-cycle arrest in glioblastoma cells. As predicted by in silico assays, miR-424 could decrease expressions of KRAS, RAF1, MAP2K1, EGFR, PDGFRA, AKT1, and mTOR. Direct interactions between miR-424-5p and RAF1 and AKT1 oncogenes has been verified by dual-luciferase reporter assay [11].
Contrary to glioblastoma cells, expression of miR-424-5p has been reported to be increased in colorectal cancer cell lines. miR-424-5p can promote proliferation and metastatic-related phenotypes through directly binding with SCN4B [12]. In laryngeal squamous cell carcinoma cells, up-regulation of miR-424-5p has enhanced proliferation, migratory aptitude, invasiveness, and adhesion of cancer cells with an important effect on cell cycle progression. In addition, CADM1 has been identified a direct target of miR-424-5p in these cells [13].
An experiment in lung cancer cells has shown that both miR-424-3p and miR-424-5p can prevent proliferation, migration, and invasiveness of these cells. In addition, miR-424-3p but not miR-424-5p could enhance chemosensitivity of lung cancer cells via influencing expression of YAP1 [14].
Different types of Small nucleolar RNA host gene (SNHG) family members have been shown to regulate expression levels of miR-424 in multiple cancers. SNHG family belongs to lncRNAs and have oncogenic roles in the malignancies [15]. For evaluation of their relation to miR-424, following studies have been conducted: In osteosarcoma cell lines Saos-2, MG63, HOS and U2OS, SNHG1 acts as a molecular sponge for miR-424-5p. After knocking down SNHG1, expression levels of miR-424-5p rises and this miRNA can target 3′-UTR of FGF2, resulting in reduced proliferation, migration and invasion [16]. The same molecular mechanism also applies for cervical cancer cells, but the difference is that sponging molecule is SNHG12 in this case and there is no FGF2 targeting [17]. Finally, in T98G and LN229 glioma cells, treatment with Ropivacaine causes SNHG16 levels to drop, and subsequent up-regulation of miR-424-5p happens, which not only suppresses proliferation and migration, but also induces apoptosis in glioma cells [18].
E2F family of TFs (transcription factors) are TFs that were firstly studied in 1987 [19]. This family can either be activators or suppressors of transcription [20]. Interestingly, E2Fs are dysregulated in a variety of cancers and it has been demonstrated that they can be regulated by miR-424. For instance, in hepatocellular carcinoma, forced expression of miR-424-5p and miR-424 is followed by downregulation of E2F7 and E2F3, respectively [21, 22]. Downregulation of these two TFs is favorable and contributes to normal-like cell properties. In cases of endometrial carcinoma, up-regulation of miR-424 diminishes E2F6 and E2F7 levels in HEC-1A, Ishikawa and HEC-1B cells, which in turn inhibits migration, invasion, and colony formation of cells [23, 24]. Lastly, in non-small cell lung cancer cell lines A549 and H460, elevation of miR-424 directly targets expression of E2F6, and consequently reduced proliferation occurs [25].
X-inactive specific transcript (XIST) belong to lncRNAs, and is famously known as X chromosome inactivator in females [26]. It is of great importance to know that this lncRNA can act as a molecular sponge for miR-424-5p in two types of cancer: firstly, in pituitary adenoma cell lines, depletion of XIST is followed by up-regulation of miR-424-5p, and as expected, reduced proliferation, migration and invasion occurs because of bFGF targeting by miR-424-5p [27]. A more detailed mechanism of XIST is demonstrated in hepatocellular carcinoma, in which inhibiting XIST expression contributes to overexpression of miR-424-5p. This miRNA degrades OGT and suppresses RAF1 glycosylation, resulting in favorable normal features in HCC cell lines [28].
Table 1 summarizes the role of miR-424 in different cancer cell lines.
Table 1.
Role of miR-424 in cancer cell lines (∆: knockdown or deletion, EMT: Epithelial mesenchymal transition, TRAIL: Tumor necrosis factor-related apoptosis-inducing ligand, 5-FU: 5- Fluorouracil, DDP: cisplatin)
| Tumor type | Targets/Regulators and signaling pathways | Cell line | Function | References |
|---|---|---|---|---|
| Neuroblastoma | DCLK1 | SK-N-SH and Be2C |
↑ miR-424 → ↓ DCLK1: ↓ invasion ↓ EMT process |
[9] |
| Oral squamous cell carcinoma | Circ_0004872 | SCC‐6, HN4, SCC‐9, CAL‐27 and SCC‐4 |
↑ circ_0004872 (which sponges miR-424-5p): ↓ proliferation ↓ invasion ↓ glycolysis ↑ apoptosis |
[29] |
| CircGDI2/SCAI | CAL-27 and SCC-15 |
↑ CircGDI2 (which sponges miR-424-5p) → ↑ SCAI: ↓ proliferation ↓ invasion ↓ migration ↓ glycolysis ↑ apoptosis |
[30] | |
| Osteosarcoma |
miR-424-5p/ VEGFA axis |
SaOS2 and MG63 |
Treatment with melatonin: ↑ miR-424-5p → ↓ VEGFA: ↓ angiogenesis |
[10] |
| Glioblastoma |
ERBB pathway: KRAS, RAF1, MAP2K1, EGFR, PDGFRA, AKT1 and mTOR |
U-251 and U-87 |
↑ miR-424-5p (which targets RAF1 & AKT1 and ERRB pathway related genes): ↓ proliferation ↓ migration ↑ apoptosis |
[11] |
| Colorectal cancer | SCN4B |
FHC and CRC cell lines (HCT8, HT29, HCT116, SW480, and SW620) |
↑ miR-424-5p → ↓ SCN4B: ↑ proliferation ↑ metastasis |
[12] |
| Src/focal adhesion kinase signaling mediated EMT | 5-fluorouracil-resistant HT-29 |
↑ miR-424-5p → ↓ Src/focal mediated EMT: ↓ resistance to 5-FU |
[31] | |
| AKT3 and PSAT1 | HCT116 and RKO |
↑ miR-424 → ↓ AKT3 & PSAT1: ↓ proliferation |
[32] | |
| TGFBR3 | Lovo |
↓ miR-424-5p → ↑ TGFBR3: ↓ proliferation ↑ apoptosis ↓ invasion ↓ migration |
[33] | |
| circTBL1XR1/Smad7 | LoVo, SW620, IEC-6, HCT 116 and SW480 |
∆ circTBL1XR1 (which sponges miR-424) → ↑ miR-424 → ↓ Smad7: ↓ proliferation ↓ invasion ↓ migration |
[34] | |
| - | SW480 |
∆ miR-424-5p: ↓ proliferation ↓ invasion ↓ migration ↓ colony formation |
[35] | |
| Colorectal cancer continued | FENDRR | HCT116, SNU-C2B, NCI-H498 and HCT-15 |
↑ FENDRR (which sponges miR-424-5p): ↓ proliferation ↓ invasion ↓ migration |
[36] |
| Laryngeal squamous cell carcinoma | CADM1 | FD-LSC-1 and TU-177 |
↑ miR-424-5p → ↓ ADM1: ↑ proliferation ↑ migration ↑ invasion |
[13] |
| Osteosarcoma | CDC25A/CCNA2 /CCNE1 | U2OS and HAL |
↑ miR-424 → ↓ CCNA2: ↓ proliferation ↓ migration ↓ cell cycle arrest |
[37] |
| SNHG1/FGF2 | Saos-2, MG63, HOS and U2OS |
∆ SNHG1 (which targets miR-424-5p) → ↑ miR-424-5p → ↓ FGF2: ↓ proliferation ↓ migration ↓ invasion |
[16] | |
| circ‐LARP4 | MG63 |
↑ circ‐LARP4 (which sponges miR-424): ↓ viability ↑ sensitivity to cisplatin and doxorubicin |
[38] | |
| Fatty acid synthase (FASN) | U2OS |
↑ miR-424 → ↓ FASN: ↓ migration ↓ invasion |
[39] | |
| LINC01116/ HMGA2 | MG-63 |
∆ LINC0116 (which silences miR-424-5p via EZH2) → ↑ miR-424-5p → ↓ HMGA2: ↓ viability ↓ migration ↓ invasion ↓ EMT process |
[40] | |
| Cutaneous squamous cell carcinoma | LINC00641 | A431 |
↑ LINC00641 → ↓ miR-424 ↓ proliferation ↓ migration ↓ invasion |
[41] |
| Cutaneous malignant melanoma | TINCR/LATS1 axis | M14, A375 and MV3 |
↑ TINCR (which sponges miR-424-5p) → ↑ LATS1: ↓ proliferation ↑ apoptosis ↓ invasion |
[42] |
| Triple negative breast cancer | PD-L1 | MDA-MB-231 |
↑ miR-424-5p → ↓ PD-L1: ↑ apoptosis |
[43] |
| Breast cancer |
PTEN/PI3K/ AKT/mTOR axis | PD-L1 |
MDA-MB-231 |
Treatment with Taxol + miR-424-5p → ↓ PTEN/PI3K/ AKT/mTOR axis and PD-L1: ↓ proliferation ↑ apoptosis ↓ colony formation ↑ cell cycle arrest |
[44] |
| Ginsenoside Rg3/ ATXN8OS/ EYA1, CHRM3, and DACH1 axis | MCF-10A, MCF-7, and MDA-MB-231 |
Treatment with Rg3 → ↓ ATXN8OS (which sponges miR-424-5p) → ↑ miR-424-5p → ↓ EYA1, CHRM3, DACH1: ↓ proliferation ↑ apoptosis |
[45] | |
| CDK1 | MDA-MB-231 and HCC1937 |
↑ miR-424-5p → ↓ CDK1: ↓ proliferation ↓ colony formation |
[46] | |
| LINC00473/CCNE1 | - |
∆ LINC00473 (which sponges miR-424-5p) → ↑ miR-424-5p → ↓ CCNE1: ↓ proliferation ↓ migration ↓ invasion ↓ EMT process |
[47] | |
|
Bax and Beclin-1 Bcl-2 and c-Myc STAT-3 and Oct-3 |
MCF-7 |
↑ miR-424-5p (in combination with miR-142-3p) → ↑Bax and Beclin-1 + ↓ Bcl-2 and c-Myc + ↓ STAT-3 and Oct-3: ↓ proliferation ↑ cell cycle arrest |
[48] | |
| DCLK1 |
DU4475, HCC1806 and MDA-MB-468 |
↑ miR-424-5p → ↓ DCLK1: ↓ proliferation ↓ motility |
[49] | |
| Renal cancer | CDC2/WEE1 | 786-O |
↑ miR-424 → ↓ WEE1 → ↑ CDC2: ↓ proliferation ↑ apoptosis |
[50] |
| Tongue squamous cell carcinoma | TGFBR3 | CAL-27 |
↑ miR-424 → ↓ TGFBR3: ↑ proliferation ↑ migration ↑ EMT |
[51] |
| Gastric cancer | ABCC2 | SGC-7901 and SGC-7901/DDP |
↓ miR-424-3p: ↑ resistance to DDP in SGC-7901 ↑ miR-424-3p: ↓ proliferation in SGC-7901/DDP |
[52] |
| Smad3/TGF-β signaling pathway | MGC803, BGC823, SGC7901, AGS and HGC27 |
↑ miR-424-5p → ↓ Smad3 → ↓ TGF-β: ↑ proliferation |
[53] | |
| MBNL1-AS1/Smad7 | AGS, MGC803, BGC-823, SGC-7901, HGC-27 |
↑ MBNL1-AS1 → ↓ miR-424-5p → ↑ Smad7: ↓ proliferation ↓ migration ↓ invasion ↑ apoptosis |
[54] | |
| Circular RNA_LARP4/LATS1 | SGC-7901, MKN-45, MKN-28, HGC-27, MGC-803, AGS and BGC-823 |
↑ LARP4 (which sponges miR-424-5p) → ↑ LATS1: ↓ proliferation ↓ invasion |
[55] | |
| Esophageal squamous cell carcinoma | SIRT4 | HEEC, EC9706, Eca-109, KYSE-150 and TE-1 |
↑ miR-424-5p → ↓ SIRT4: ↑ proliferation |
[56] |
| Smad7 | EC9706, Eca109, EC-1 |
↑ miR-424-5p → ↓ Smad7: ↓ proliferation ↓ invasion ↓ EMT process |
[57] | |
| Acute myeloid leukemia | PLAG1 | HL-60, NB4, HL-60/ADM, K562 |
↑ miR-424 → ↓ PLAG1: ↑ sensitivity to TRAIL |
[58] |
| Prostate cancer |
ESE3/EHF /COP1/STAT3 |
DU145 |
↓ ESE/EHF → ↑ miR-424-5p → ↓ COP1 (which degrades STAT3) → ↑ STAT3: ↑ proliferation ↑ migration |
[59] |
| Infantile hemangioma | VEGFA | XPTS-1 |
Treatment with propranolol: ↑ miR-424 → ↓ VEGFA: ↓ viability ↓ invasion ↑ apoptosis |
[60] |
| Pituitary adenoma | JAG1 | GH3 |
↑ miR-424-3p → ↓ JAG1: ↓ proliferation |
[61] |
| XIST/bFGF axis |
∆ XIST (which sponges miR-424-5p) → ↑ miR-424-5p → ↓ bFGF: ↓ proliferation ↓ migration ↓ invasion ↑ apoptosis |
[27] | ||
| Cervical cancer | KDM5A/Suz12 | CaSki |
Human papillomavirus type 16 E7 induction: ↑ KDM5A (which binds to promoter region of miR-424-5p and inhibits it) → ↑ Suz12: ↑ proliferation ↑ invasion |
[62] |
| SNHG12 | C33A, ME-180, CaSki, HeLa and SiHa |
∆ SNHG12 (which sponges miR-424-5p) → ↑ miR-424-5p: ↓ proliferation ↓ migration ↓ invasion |
[17] | |
| Head and Neck Squamous Cell Carcinoma | LAMC1/ Wnt/β-catenin signaling pathway | HUVEC |
↑ miR-424-5p → ↓ LACM1 → ↓ Wnt/β-catenin signaling pathway: ↓ angiogenesis ↓ migration |
[63] |
| Hepatocellular carcinoma | E2F7 | HB-8064, HB-8065, CRL2235, CRL-2237 and THLE-3 |
↑ miR-424-5p → ↓ E2F7: ↓ proliferation ↑ cell cycle arrest |
[21] |
| KIF2A | Huh7 and HepG2 |
↑ miR-424-5p (which is epigenetically suppressed) → ↓ KIF2A: ↓ EMT process ↓ proliferation ↑ apoptosis |
[64] | |
| XIST/OGT/RAF1 | MHCC97L, MHCC97H, SNU-398, SMMC7221, and Huh7 |
↓ XIST (which sponges miR-424-5p) → ↑ miR-424-5p → ↓ OGT → ↑ RAF1: ↓ EMT process ↓ proliferation |
[28] | |
| ATG14 | HepG2, SMMC-7721, Huh-7, MHCC97H and HCCLM3 |
↑ miR-424-5p → ↓ ATG14: ↓ proliferation ↑ apoptosis ↓ autophagy |
[65] | |
| - | SMMC-7721, Huh-7, HepG2, Bel-7402, and SK-HEP-1 |
↑ miR-424: ↓ proliferation ↓ migration ↓ invasion |
[66] | |
| CircCBFB/ATG14 | Huh-7 and HCCLM3 |
↑ CircCBFB (which sponges miR-424-5p) → ↑ ATG14: ↑ proliferation ↑ autophagy |
[67] | |
| CDKN2B-AS1 | Huh7, Hep3B and Sk-Hep1 |
↓ CDKN2B-AS1 (which sponges miR-424-5p) → ↑ miR-424-5p: ↓ proliferation ↓ migration ↓ invasion ↓ EMT process |
[68] | |
| Hepatocellular carcinoma continued | TRIM29 | MHCC-97H, HepG2, SMMC-7721, and Huh-7 |
↑ miR-424-5p → ↓ TRIM29: ↓ proliferation ↓ migration ↓ invasion ↓ colony formation |
[69] |
| Akt3/E2F3 axis | SMMC7721, HepG2, HUH7, MHCC97-L, MHCC97-H and HCCLM3 |
↑ miR-424 → ↓ Akt3/E2F3: ↓ proliferation ↓ colony formation |
[22] | |
| LINC00922/ARK5 axis | SNU-182 and SK-Hep1 |
↑ LIN C00922 → ↓ miR-424-5p → ↑ ARK5: ↑ viability ↑ migration ↑ invasion ↑ EMT process |
[70] | |
| DLX6-AS1/WEE1 | SK‐HEP‐1 and Hep3B |
↑ DLX6-AS1 (which sponges miR-424-5p) → ↑ WEE1: ↑ proliferation ↑migration ↑ invasion |
[71] | |
| Cholangiocarcinoma | LINC00665/BCL9L | HuCCT1-Gem and SNU-245-Gem |
∆ LINC00665 (which sponges miR-424-5p) → ↑ miR-424-5p → ↓ BCL9L: ↑ apoptosis ↓ EMT process ↓ resistance to gemcitabine |
[72] |
| Intrahepatic cholangiocarcinoma | ARK5 | CLP-1, RBE and HuCCT-1 |
↑ miR-424-5p → ↓ ARK5: ↓ migration ↓ invasion |
[73] |
| Bladder cancer | DNMT1/ EGFR signaling | HT1197 and HT1376 |
↑ DNMT1 (which suppresses miR-424 expression): ↑ EGFR signaling: ↑ proliferation ↑ migration ↑ EMT process |
[74] |
| LINC00355/ HMGA2 |
T24, HT-1197, SW780, HT-1376, UM-UC-3, TCCSUP, KU1919, and VMCUB1 |
↑ LINC00355 (which sponges miR-424-5p) → ↑ HMGA2: ↑ migration ↑ invasion ↑ EMT process |
[75] | |
| Endometrial carcinoma | MMSET | Ishikawa and HEC-1 |
↑ miR424 → ↓ MMSET: ↓ invasion ↓ sphere formation |
[76] |
| IGF-1R | HEC‐1A, HEC‐1B, AN3CA, and Ishikawa |
↑ miR-424 → ↓ IGF‐1R: ↓ viability ↓ proliferation ↓ EMT process |
[77] | |
| E2F6 | HEC-1A, Ishikawa |
↑ miR-424-3p → ↓ E2F6: ↓ migration ↓ invasion ↓ EMT process |
[23] | |
| E2F7 | Ishikawa and HEC-1B |
↑ miR-424 → ↓ E2F7: ↓ proliferation ↓ colony formation ↑ apoptosis |
[24] | |
| SPTBN2 /PI3K/AKT | AN3C and Ishikawa |
↑ miR-424-5p → ↓ SPTBN2 → ↓ PI3K/AKT: ↓ proliferation ↓ colony formation |
[78] | |
| Ovarian cancer/epithelial ovarian cancer (EOC) | LGALS3 | SK-OV-3 and TOV-21G |
↑ miR-424-3p → ↓ LGALS3: ↑ sensitivity to cisplatin |
[79] |
| NANOG/WEE1 | SKOV3, OVCAR3, OVCAR5, and OVCAR8 |
↑ miR-424 (which is suppressed by NANOG) → ↓ WEE1: ↓ proliferation ↓ migration ↓ colony formation ↑ sensitivity to carboplatin |
[80] | |
| MYB | SKOV-3, HO8910 and A2780 |
↑ miR-424-5p → ↓ MYB: ↓ proliferation ↓ migration ↓ invasion |
[81] | |
| CCNE1 | SKOV3, HO8910 and A2780 |
↑ miR-424-5p → ↓ CCNE1: ↓ proliferation ↑ cell cycle arrest |
[82] | |
| ACSL4 |
HO8910 and SKOV3 |
↑ miR-424-5p → ↓ ACSL4: ↓ ferroptosis |
[83] | |
| Ovarian clear cell carcinoma | DCLK1 |
ES-2 and RMG-1 |
↑ miR-424 → ↓ DCLK1: ↓ migration ↓ invasion ↓ EMT process |
[84] |
| Pancreatic ductal adenocarcinoma (PDAC) | SOCS6 | PANC-1, AsPC-1, BxPC-3 and MIAPaCa-2 |
↓ miR-424-5p → ↑ SOCS6: ↓ proliferation ↓ migration ↓ invasion ↑ apoptosis |
[85] |
| Glioma | CCAT2/VEGFA | A172, U251 and HCMEC/D3 |
↑ miR-424 (which is sponged by CCAT2) → ↓ VEGFA: ↓ Angiogenesis |
[86] |
| GAS5/PRC2 | LN229, A172, U373, SHG44 and NHA |
↑ GAS5 (which interacts with EZH2 in PRC2) → ↓ methylation of miR-424 promoter → ↑ miR-424: ↑ apoptosis ↓ proliferation ↓ migration ↓ invasion |
[87] | |
| KIF23 | A172, SHG-44, T98, LN18, and LN229 |
↑ miR-424 → ↓ KIF23: ↓ EMT process |
[88] | |
|
FAM87A/PPM1H PI3K/Akt Signaling Pathway |
T98G, A172, SNB19 and U251 |
↑ FAM87A (which sponges miR-424-5p) → ↑ PPM1H → ↓ PI3K/Akt Signaling Pathway: ↓ viability ↓ migration ↓ invasion |
[89] | |
| SNHG16 | T98G and LN229 |
Treatment with Ropivacaine: ↓ SNHG16 (which targets miR-424-5p) → ↑ miR-424-5p: ↑ apoptosis ↓ proliferation ↓ migration ↓ invasion |
[18] | |
| Nasopharyngeal carcinoma | AKT3 | NPC-1 |
↑ miR-424-5p → ↓ AKT3: ↓ proliferation ↓ migration ↓ invasion |
[90] |
| Non-small cell lung cancer | YAP1 |
H827, H2172, H441, A549, H1975, and PC14 |
∆ miR-424-3p (which targets YAP1): ↑ proliferation ↑ migration ↑ invasion ↑ resistance to Paclitaxel |
[14] |
| PTEN/PI3K/Akt pathway | A549 and H460 |
Treatment with baicalein: ↓ miR‐424‐3p → ↑ PTEN → ↓ PI3K/AKT: ↑ cisplatin sensitivity ↓ cell survival |
[91] | |
| E2F6 | A549 and H460 |
↑ miR-424 → ↓ E2F6: ↓ proliferation ↓ invasion |
[25] | |
| – | H596 and SW900 |
↓ miR-424: ↓ proliferation ↑ cell cycle arrest |
[92] |
Animal models
Animal models of different types of cancers, including mammary tumors, aggressive osteosarcoma, gastric cancer, esophageal squamous cell carcinoma, thyroid cancer and glioma have been established to assess the impact miR-424 dysregulation on the tumor burden (Table 2). In gastric cancer xenograft models, the results of two studies are contradictory. While up-regulation of miR-424-3p has led to reduction of tumor growth and metastasis in one study [52], another study has reported that over-expression of mir-424-5p has the opposite effect [53]. In esophageal squamous cell carcinoma, both conducted studies have confirmed an oncogenic role for this miR-424 [56, 93]. The results of other studies in xenograft models are shown in Table 2.
Table 2.
Impact of miR-424 in carcinogenesis in vivo (∆ knockdown or deletion, SCID: severe combined immunodeficiency, SPF: specific pathogen-free)
| Tumor type | Animal models | Results | References |
|---|---|---|---|
| Osteosarcoma | SCID mice |
↑miR-424: ↓tumor growth |
[37] |
| Gastric cancer | BALB/c nude mice |
↑ miR-424-3p: ↓ tumor growth ↓ metastasis |
[52] |
| BALB/c nude mice |
↑ mir-424-5p: ↑ tumor size |
[53] | |
| Esophageal squamous cell carcinoma | BALB/c-nude mice |
∆ miR-424-5p: ↓tumor growth |
[56] |
| BALB/c nude mice | ↑ miR-424: tumorigenesis | [93] | |
| Thyroid cancer | BALB/c nude mice |
∆ miR-424-5p: ↓ metastasis | ↑ miR-424-5p: ↑ Lung metastasis |
[94] |
| Ovarian cancer | Nude mice |
↑ miR-424-5p ↓ tumor growth ↓ angiogenesis |
[81] |
| Hepatocellular carcinoma | Nude mice |
↑ miR-424-5p: ↓ tumor growth |
[64] |
| SPF BALB/c nude mice |
↓ XIST: ↑ miR-424-5p: ↓ tumor growth |
[28] | |
| BALB/c nude mice |
↑ miR-424-5p: ↓ tumor growth |
[69] | |
| Nude mice |
↑ miR-424-5p: ↓ tumor growth |
[66] | |
| Nude mice |
↑ miR-424-5p: ↓ tumor growth |
[22] | |
| Colorectal cancer | BALB/c nude mice |
↓ miR-424-5p ↓ tumor growth |
[33] |
| Glioma | BALB/c nude mice |
↑ miR-424: ↓ tumor growth |
[88] |
Assays in clinical samples
Expression of miR-424 has been evaluated in a variety of malignant tissues. Experiments in ovarian cancer, cervical cancer, hepatocellular carcinoma, neuroblastoma, breast cancer, osteosarcoma, intrahepatic cholangiocarcinoma, prostate cancer, endometrioid endometrial cancer, non-small cell lung cancer, hemangioma and gastric cancer have reported down-regulation of miR-424. On the other hand, this miRNA has been found to be up-regulated in melanoma, laryngeal and esophageal squamous cell carcinomas, glioma, multiple myeloma and thyroid cancer. In colorectal cancer, both patterns have been reported (Table 3). In neuroblastoma tissues, down-regulation of miR-424 has been found to be accompanied by up-regulation of its target gene DCLK1 [9]. On the other hand, miR-424-5p has been found to be upregulated in laryngeal squamous cell carcinoma samples versus adjacent normal margin tissues. Over-expression of miR-424-5p has been associated with poor differentiation, advanced tumor stages and cervical lymph node involvement. In silico analyses have shown that target genes of this miRNA are principally enriched in cell cycle, cell division, and negative regulation of cell migration [13]. In patients with cervical cancer, down-regulation of miR-424 has been reported to be associated with low level of differentiation of cancer cells, advanced clinical stages and metastasis to lymph nodes [95]. Furthermore, in patients with non-small cell lung cancer, low levels of miR-424-3p have been associated with disease progression and overall prognosis [14].
Table 3.
Dysregulation of miR-424 in clinical specimens (O-S: overall survival, DFS: disease-free survival, ANT: adjacent normal tissue, CFFS: Clinical failure-free survival, TCGA: the cancer genome atlas, AFP: Alpha-fetoprotein, HBV: hepatitis B virus, HPV: human papilloma virus, RFS: recurrence-free survival)
| Tumor type | Samples | microRNA Type | Expression (tumor vs normal) | Kaplan–Meier analysis (impact of miR-424) | Association of miR-424 expression with clinicopathologic characteristics | References |
|---|---|---|---|---|---|---|
| Ovarian cancer (OC)/ epithelial ovarian cancer (EOC) | 38 OC + paired ANT | 5p | Downregulated | – | – | [83] |
| 31 EOC + paired ANT | miR-424 | Downregulated | Lower O-S and DFS | – | [96] | |
| 85 OC + 43 paired ANT | 5p | Downregulated | – | – | [81] | |
| 83 EOC + 19 paired ANT | 5p | Downregulated | Shorter O-S and RFS | Associated with poor differentiation, advanced FIGO stage, residual tumor size and lymph node metastasis | [82] | |
| Ovarian clear cell carcinoma (OCCC) | 30 OCCC + paired ANT | miR-424 | Downregulated | – | – | [84] |
| Cervical cancer (CC) | 30 CC + paired ANT | miR-424 | Downregulated | Poor O-S | – | [99] |
| 147 CC + 74 normal tissues | miR-424 | Downregulated | Poor O-S | Associated with tumor differentiation, advanced clinical stages and lymph node metastases | [95] | |
| 42 HPV16 positive + 17 HPV16 negative + 13 normal tissues | 5p | Downregulated (especially in HPV16 positive) | – | – | [62] | |
| Hepatocellular carcinoma (HCC) | 156 HCC + paired ANT | 5p | Downregulated | – | – | [100] |
| 60 HCC + paired ANT | miR-424 | Downregulated | Poor O-S | Associated with advanced clinical stage | [101] | |
| 127 HCC + paired ANT | miR-424 | Downregulated | Poor O-S | Associated with Lymph node metastasis, vascular invasion, and clinical stage | [102] | |
| 30 HCC + paired ANT | 5p | Downregulated | – | – | [64] | |
| 80 HCC + paired ANT | 5p | Downregulated | – | – | [28] | |
| 36 HCC + paired ANT | 5p | Downregulated | Poor survival rate | Associated with tumor size, HBV infection, AFP content, and TNM | [65] | |
| 90 HCC + paired ANT | 5p | Downregulated | Poor O-S | Associated with AFP, TNM stage and intrahepatic metastasis | [69] | |
| 121 HCC + paired ANT | miR-424 | Downregulated | – | Associated with recurrence | [66] | |
| 96 HCC + paired ANT | miR-424 | Downregulated | Poor O-S | Associated with tumor size, Tumor nodule number, TNM stage and BCLC stage | [22] | |
| 50 HCC + paired ANT | 5p | Downregulated | – | – | [103[ | |
| 40 HCC + paired ANT | 5p | Downregulated | – | – | [70] | |
| Melanoma | Serum and tissue of melanoma patients | miR-424 | Upregulated | Lower O-S and DFS | Associated with tumor thickness, metastasis and tumor stage and ulceration | [104] |
| Cutaneous malignant melanoma (CMM) | 60 CMM + paired ANT | 5p | Downregulated | – | Associated with advanced TNM stage | [42] |
| Neuroblastoma | 49 neuroblastoma tissues + paired ANT | miR-424 | Downregulated | – | – | [9] |
| Breast cancer | TCGA database | miR-424 | Downregulated | Poor O-S | Associated with high-grade, larger tumor size, triple-negative status, stronger cell proliferation, and GGI signature | [105] |
| 17 BC + paired ANT | 5p | Downregulated | Poor O-S | – | [46] | |
| 84 BC + 20 paired ANT | 5p | Downregulated | – | Associated with clinical stage, larger tumor size, lymph node metastasis and distant metastasis | [49] | |
| Colorectal cancer (CRC) | GSE108153 (21 CRC + paired ANT) + TCGA database | 5p | Upregulated | – | – | [12] |
| 24 CRC + paired ANT | miR-424 | Downregulated | – | – | [106] | |
| 65 CRC + paired ANT | 5p | Upregulated | – | Associated with tumor differentiation, tumor infiltration depth, TNM stage, vascular invasion, lymph node metastasis and distant metastasis | [33] | |
| 59 CRC + paired ANT | 5p | Upregulated | – | Associated with Dukes’ stage, depth of invasion and pathological type | [35] | |
| 20 CRC + paired ANT | 5p | Upregulated | – | – | [36] | |
| Laryngeal squamous cell carcinoma (LSCC) | 106 LSCC + paired ANT | 5p | Upregulated | Poor O-S | Associated with advanced T stage and lymph node metastasis | [13] |
| Nasopharyngeal carcinoma (NPC) | 40 NPC + 26 healthy controls skin (obtained from plastic surgery) | 5p | Downregulated | – | Associated with TNM stage and lymph node metastasis | [90] |
| Oral squamous cell carcinoma (OSCC) | Saliva of 43 OSCC + 44 healthy controls | 3p | Downregulated | – | – | [107] |
| 60 OSCC + paired ANT | 5p | Upregulated | – | – | [29] | |
| Blood sample of 15 OSCC + 15 healthy controls | 5p | Upregulated | – | – | [97] | |
| 30 OSCC + paired ANT | 5p | Upregulated | – | – | [30] | |
| Osteosarcoma (OS) | Plasma of 20 OS + 15 healthy controls | miR-424 | Downregulated | – | – | [37] |
| 61 OS + paired ANT | 5p | Downregulated | – | – | [16] | |
| Intrahepatic cholangiocarcinoma (ICC) | 19 ICC + paired ANT | 5p | Downregulated | – | – | [73] |
| Prostate cancer (PCa) | 535 PCa tissues | 3p | Downregulated | Poor CFFS | Associated with aggressiveness of disease | [108] |
| 48 PCa + 21 healthy controls tissue | 5p | Upregulated | – | – | [59] | |
| Endometrioid Endometrial Cancer (EEC)/Endometrial carcinoma (EC) | 24 EEC + paired ANT | 5p | Downregulated | – | – | [78] |
| 50 EC + 10 fibroid samples as controls | miR-424 | Downregulated | – | – | [77] | |
| 32 EC + paired ANT | 3p | Downregulated | – | Associated with clinical stage and lymph node metastasis | [23] | |
| 11 EC + paired ANT | miR-424 | Downregulated | – | – | [24] | |
| Non-small cell lung cancer (NSCLC) | 90 NSCLC + paired ANT | 5p | Downregulated | Poor O-S | Associated with clinical stage, lymph node metastasis, differentiation degree, and tumor volume | [14] |
| 233 NSCLC + paired ANT | miR-424 | Upregulated in advanced stage (no difference with normal tissues) | Poor O-S | Associated with advanced clinical stage, TNM stage and lymph node metastasis | [92] | |
| Glioma | 76 glioma tissues + paired ANT | 5p | Upregulated | Poor O-S | – | [89] |
| 54 glioma tissues + 12 healthy controls tissues (obtained from traumatic brain injury) | miR-424 | Downregulated | Poor O-S | Associated with WHO grade and KPS | [88] | |
| Esophageal squamous cell carcinoma cell (ESCC) | 30 ESCC + 10 healthy controls | miR-424 | Upregulated | Poor O-S | – | [93] |
| 32 ESCC + paired ANT | 5p | Downregulated | – | Associate with differentiation and lymph node metastasis | [57] | |
| Hemangioma (HM) | 7 Senile HM + 3 venous malformations + 4 angiosarcoma + 4 venous lakes + 3 infantile HM | miR-424 | Downregulated | – | Associate with Abnormal Angiogenesis | [109] |
| Infantile hemangioma (IH) | 16 IH + 16 normal subcutaneous tissues | miR-424 | Downregulated | – | – | [110] |
| 13 IH + paired ANT | miR-424 | Downregulated | – | – | [60] | |
| Gastric cancer (GC) | 48 GC + paired ANT | miR-424 | Downregulated | Poor O-S | Associated with TNM stage and lymph node metastasis | [98] |
| 60 GC + paired ANT | 5p | Upregulated | – | – | [54] | |
| TCGA database (387 GC + 41 paired ANT) | 5p | Upregulated | Recurrence of GC | – | [55] | |
| Pancreatic ductal adenocarcinoma (PDAC) | 24 PDAC + paired ANT | 5p | Upregulated | – | – | [85] |
| Multiple myeloma (MM) | Serum of 81 MM patients + 50 healthy controls | miR-424 | Upregulated | Associated with clinical stage | [111] | |
| Thyroid cancer (TC) | TCGA dataset + 10 TC + paired ANT | 5p | Upregulated | – | Associated with distant metastasis | [94] |
Out of 4 different studies conducted on ovarian cancer/epithelial ovarian cancer patients, two have reported downregulation of miR-424 and other two verified downregulation of miR-424-5p in tumor tissues compared with adjacent normal tissues [81–83, 96].
In Oral squamous cell carcinoma (OSCC), three different studies confirmed up-regulation of miR-424-5p both in OSCC tissues and blood samples [29, 30, 97]. Discordant to mentioned studies, miR-424-3p has shown to be under-expressed in saliva of OSCC patients, and its levels can be used as a diagnostic marker to differentiate OSCC patients and healthy controls, with an AUC value of 0.732, sensitivity of 0.605 and 0.818 specificity.
It is worth mentioning that a great number of research on clinical specimens have been conducted in hepatocellular carcinoma (HCC) patients. In total, after evaluating expression of miR-424 and miR-424-5p in 886 pairs of HCC tissues in 11 independent studies, it is concluded that both of these miRNAs are downregulated in HCC tissues. According to Kaplan–Meier analysis, downregulated levels of miR-424-5p is associated with shorter over-all survival in HCC patients [65]. In addition, diminished levels of this miRNA is associated with alpha-fetoprotein content and HBV infection in HCC [65].
In gastric cancer (GC) patients, miR-424 has been verified to be downregulated [98] but miR-424-5p is upregulated in tumor expression profile [54, 55]. In case of miR-424 downregulation, NNT-AS1 acts as a molecular sponge and inhibits miR-424 and this phenomenon is accompanied by activation of E2F1, a transforming transcription factor [98].
Associations between dysregulation of miR-424 and clinical outcome have been demonstrated in ovarian, cervical, breast, prostate, lung, melanoma, colorectal and other cancers (Table 3).
In patients with colorectal cancer, cancer cells have been shown to release miR-424-5p into peripheral blood in the form of exosomes. Notably circulating exosomal levels of miR-424-5p can separate patients with early stage of colorectal cancer from healthy individuals with area under the ROC curve (AUC) value of 0.82 [12]. In patients with multiple myeloma, serum levels of this miRNA could be used as a diagnostic marker with AUC value of 0.95 [111]. Diagnostic role of miR-424 has also been evaluated in hepatocellular carcinoma, prostate cancer and renal cell carcinoma revealing different AUC values (Table 4).
Table 4.
Diagnostic value of miR-424 in cancers (BPH: benign prostate hyperplasia)
| Tumor type | Samples | Distinguish between | Area under curve | Sensitivity (%) | Specificity (%) | References |
|---|---|---|---|---|---|---|
| Hepatocellular carcinoma (HCC) | Serum of HCC patients and healthy controls | HCC vs healthy controls | 0.768 | 75.0% | 72.4% | [112] |
| Multiple myeloma (MM) | 81 MM + 50 healthy controls | MM vs healthy controls | 0.952 | 95.0% | 87.2% | [111] |
| Prostate cancer (PC) | Serum of 36 PCa + 54 BPH | PCa vs BPH | 0.671 | 31.94% | 94.87% | [113] |
| Oral squamous cell carcinoma (OSCC) | Saliva of 43 OSCC + 44 healthy controls | OSCC vs healthy controls | 0.732 | 0.605 | 0.818 | [107] |
| Renal cell carcinoma (RCC) | Serum of 22 RCC + 16 healthy controls | RCC vs healthy controls | 0.7727 | 81.8% | 75.0% | [114] |
Discussion
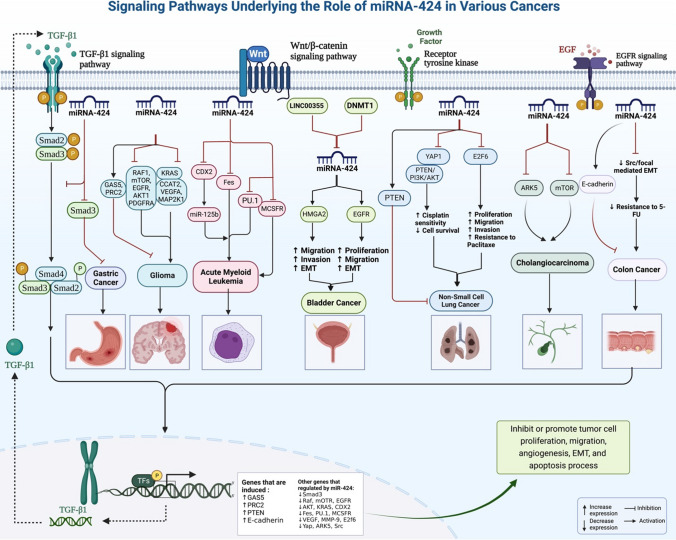
miR-424 is an example of miRNAs with crucial roles in the carcinogenesis. However, its role in this process may depend on the type of tissue, since in some tissues it makes cancer cells grow and in others it prevents cancer cells from growing. Moreover, it is possible that miR-424-3p and miR-424-5p exert different roles in the process of carcinogenesis in some cases (115) (Fig. 1). This is mostly related with the specific targets of miR-424 in each tissue. The interactions between miR-424 and tissue specific transcription factors might also been involved in the specificity of miR-424 functions in each tissue.
Fig. 1.
miRNA-424 can either inhibit or stimulate cancer cell growth depending on the context. For instance, overexpression of miR-424 can inhibit glioblastoma cell growth and migration. In addition, miR-424 has been demonstrated to induce cell-cycle arrest and enhance apoptosis in glioblastoma cells. KRAS, RAF1, MAP2K1, EGFR, PDGFRA, AKT1, and mTOR expressions may all be suppressed by miR-424, as suggested by in silico experiments
Expression of this miRNA is regulated by methylation status of its promoter and lncRNAs that sponge this miRNA [64]. LINC00641 [115], CCAT2 [116], PVT1 [117], LINC00511 [102] and NNT-AS1 [98] are among lncRNAs that act as molecular sponges for miR-424. The interactions between lncRNAs and miRNAs forms a huge and complex regulation network for regulation of gene expression at transcriptional, post-transcriptional, and post-translational levels [118]. This multilevel regulatory network is involved in the carcinogenesis. miR-424/lncRNA axes might represent potential targets for design of novel therapeutics for cancers. However, the function of these axes should be individually assessed in each type of cancer.
miR-424 has a regulatory role on activity of VEGFA, ERBB, mTOR, TGF-β and PTEN/PI3K/AKT pathways. Since miR-424 can influence expression of VEGFA, it has a pivotal role in the regulation of tumor angiogenesis [119]. Moreover, it has been shown to influence EMT process through inhibiting mTOR phosphorylation [73]. miR-424 can affect cell-cycle transition and cell apoptosis. KRAS, RAF1, MAP2K1, EGFR, PDGFRA, AKT1, and mTOR expressions can also been affected by miR-424 [11]. Therefore, a wide array cancer-related genes, pathways and cellular functions are controlled by this miRNA.
Serum levels of miR-424 can distinguish cancer patients from healthy controls with variable diagnostic power values. Diagnostic role of miR-424 has been evaluated in hepatocellular carcinoma [112], multiple myeloma [120], prostate cancer [121], oral squamous cell carcinoma [107], renal cell carcinoma [50] with the best values being obtained in multiple myeloma. Confirmation of these results in larger sample sizes from different types of cancer can broaden application of this miRNA in non-invasive methods for cancer detection. Since early diagnosis of cancer is the most efficient way to increase survival of patients, these findings have significance in this regard. Moreover, serum levels of miR-424 can be used for follow-up of patients with different types of cancers after conduction of anti-cancer therapies to detect cancer recurrence.
The association between expression levels of miR-424 and clinical outcome of patients further highlights the potential of this miRNA as a predictive biomarker in cancer patients. However, since miR-424 has opposite roles in different cancers [13], the patterns and direction of these associations depends on the role of miR-424 in each type of cancer. This note should also be considered when designing miR-424-targeting strategies in the treatment of cancer. Although there are ongoing clinical trials in phase 1 and 2 regarding their therapeutic application, there is no FDA approved miRNA-based drug in the market [122]. However, patisiran, and givosiran are two FDA approved siRNA-based drugs [123] and because of the similarity in the mechanism of action, we can anticipate miRNA-based drugs in the near future. In the cases of miR-424, the wide range of molecules being affected by this miRNA enhances the efficacy of targeted therapies in the field of cancer. However, this feature also increases the possibility of unwanted side effects.
Since the majority of studies, especially cell line studies are conducted on colorectal cancer (7 studies), osteosarcoma (5 studies), breast cancer (7 studies), gastric cancer (4 studies), hepatocellular carcinoma (11 studies), endometrial carcinoma (5 studies), ovarian cancer (6 studies), glioma (5 studies) and non-small cell lung cancer (4 studies), it might be wise to be more focused on these conditions when considering therapeutic approaches of miR-424.
Finally, miR-424 affects response of cells to 5-flurouracil [31], paclitaxel [14], gemcitabine [72] and cisplatin [52]. Thus, dysregulation of expression of miR-424 might be involved in the chemoresistance phenotype.
In brief, miR-424 is an example of miRNAs with tissue-specific impacts in the carcinogenesis. Experiments in cancer cell lines and animal models of cancer have shown feasibility and efficacy of miR-424-targeting strategies in decreasing invasiveness of cancer cells and tumor burden, respectively. Applicability of these strategies in clinical setting has not been evaluated yet. Future studies are needed to elaborate this aspect.
Acknowledgements
The authors would like to thank the clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Author contributions
SGF wrote the draft and revised it. MT designed and supervised the study. BMH, AA and NAD collected the data and designed the figure and tables.
Funding
Open Access funding enabled and organized by Projekt DEAL. Not applicable.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare they have no conflict of interest.
Ethical approval and consent to participant
Not applicable.
Consent of publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Taheri, Email: mohammad.taheri@uni-jena.de.
Nader Akbari Dilmaghani, Email: nadakbari@sbmu.ac.ir.
References
- 1.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: a signature for cancer progression. Biomed Pharmacother. 2021;138:111528. doi: 10.1016/j.biopha.2021.111528. [DOI] [PubMed] [Google Scholar]
- 3.Hussen BM, Abdullah ST, Rasul MF, Salihi A, Ghafouri-Fard S, Hidayat HJ, et al. MicroRNAs: important players in breast cancer angiogenesis and therapeutic targets. Front Mol Biosci. 2021;8:764025. doi: 10.3389/fmolb.2021.764025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghafouri-Fard S, Honarmand Tamizkar K, Hussen BM, Taheri M. MicroRNA signature in liver cancer. Pathology - Research and Practice. 2021;219:153369. doi: 10.1016/j.prp.2021.153369. [DOI] [PubMed] [Google Scholar]
- 5.Ghafouri-Fard S, Hussen BM, Badrlou E, Abak A, Taheri M. MicroRNAs as important contributors in the pathogenesis of colorectal cancer. Biomed Pharmacother. 2021;140:111759. doi: 10.1016/j.biopha.2021.111759. [DOI] [PubMed] [Google Scholar]
- 6.Hussen BM, Hidayat HJ, Ghafouri-Fard S. Identification of expression of CCND1-related lncRNAs in breast cancer. Pathology - Research and Practice. 2022;236:154009. doi: 10.1016/j.prp.2022.154009. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10a):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan MF, Yang N, Qu NY, Pan YY, Shan YQ, Li P. MiR-424 suppressed viability and invasion by targeting to the DCLK1 in neuroblastoma. Eur Rev Med Pharmacol Sci. 2020;24(10):5526–5533. doi: 10.26355/eurrev_202005_21338. [DOI] [PubMed] [Google Scholar]
- 10.Vimalraj S, Saravanan S, Raghunandhakumar S, Anuradha D. Melatonin regulates tumor angiogenesis via miR-424-5p/VEGFA signaling pathway in osteosarcoma. Life Sci. 2020;256:118011. doi: 10.1016/j.lfs.2020.118011. [DOI] [PubMed] [Google Scholar]
- 11.Gheidari F, Arefian E, Adegani FJ, Kalhori MR, Seyedjafari E, Kabiri M, et al. miR-424 induces apoptosis in glioblastoma cells and targets AKT1 and RAF1 oncogenes from the ERBB signaling pathway. Eur J Pharmacol. 2021;906:174273. doi: 10.1016/j.ejphar.2021.174273. [DOI] [PubMed] [Google Scholar]
- 12.Dai W, Zhou J, Wang H, Zhang M, Yang X, Song W. miR-424-5p promotes the proliferation and metastasis of colorectal cancer by directly targeting SCN4B. Pathol Res Pract. 2020;216(1):152731. doi: 10.1016/j.prp.2019.152731. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Liu J, Hu W, Zhang Y, Sang J, Li H, et al. miR-424-5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. 2019;12:10441–10453. doi: 10.2147/OTT.S224325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Zeng J, Zhao Z, Liu Z. Loss of MiR-424-3p, not miR-424-5p, confers chemoresistance through targeting YAP1 in non-small cell lung cancer. Mol Carcinog. 2017;56(3):821–832. doi: 10.1002/mc.22536. [DOI] [PubMed] [Google Scholar]
- 15.Zimta AA, Tigu AB, Braicu C, Stefan C, Ionescu C, Berindan-Neagoe I. An emerging class of long non-coding RNA with oncogenic role arises from the snoRNA host genes. Front Oncol. 2020;10:389. doi: 10.3389/fonc.2020.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Wang X, Liang S. Long non-coding RNA small nucleolar RNA host gene 1 knockdown suppresses the proliferation, migration and invasion of osteosarcoma cells by regulating microRNA-424-5p/FGF2 in vitro. Exp Ther Med. 2021;21(4):325. doi: 10.3892/etm.2021.9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for MiR-424-5p. Cell Physiol Biochem. 2018;45(5):2086–2094. doi: 10.1159/000488045. [DOI] [PubMed] [Google Scholar]
- 18.Liu R, Wu M, Xu G, Ju L, Xiao J, Zhong W, et al. Ropivacaine inhibits proliferation, migration, and invasion while inducing apoptosis of glioma cells by regulating the SNHG16/miR-424-5p axis. Open Life Sci. 2020;15(1):988–999. doi: 10.1515/biol-2020-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee AS, Reichel R, Kovesdi I, Nevins JR. Promoter interaction of the E1A-inducible factor E2F and its potential role in the formation of a multi-component complex. Embo j. 1987;6(7):2061–2068. doi: 10.1002/j.1460-2075.1987.tb02471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie D, Pei Q, Li J, Wan X, Ye T. Emerging role of E2F family in cancer stem cells. Front Oncol. 2021;11:723137. doi: 10.3389/fonc.2021.723137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Zhu C, Chang Q, Peng P, Yang J, Liu C, et al. MiR-424-5p regulates cell cycle and inhibits proliferation of hepatocellular carcinoma cells by targeting E2F7. PLoS One. 2020;15(11):e0242179. doi: 10.1371/journal.pone.0242179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Zheng W, Shuai X, Chang RM, Yu L, Fang F, et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6(29):27736–27750. doi: 10.18632/oncotarget.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, Nian Z, Jingjing Z, Tao L, Quan L. MicroRNA-424/E2F6 feedback loop modulates cell invasion, migration and EMT in endometrial carcinoma. Oncotarget. 2017;8(69):114281–114291. doi: 10.18632/oncotarget.23218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Qiu XM, Li QH, Wang XY, Li L, Xu M, et al. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol Rep. 2015;33(5):2354–2360. doi: 10.3892/or.2015.3812. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Lan H, Zhang M, An N, Yu R, He Y, et al. Effects of miR-424 on proliferation and migration abilities in non-small cell lung cancer A549 cells and its molecular mechanism. Zhongguo Fei Ai Za Zhi. 2016;19(9):571–576. doi: 10.3779/j.issn.1009-3419.2016.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130(2):223–236. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou K, Li S, Du G, Fan Y, Wu P, Sun H, et al. LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. Onco Targets Ther. 2019;12:7095–7109. doi: 10.2147/OTT.S208329. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Ning D, Chen J, Du P, Liu Q, Cheng Q, Li X, et al. The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. Liver Int. 2021;41(8):1933–1944. doi: 10.1111/liv.14904. [DOI] [PubMed] [Google Scholar]
- 29.Dai Y, Zhu Y, Xu H. circ_0004872 inhibits proliferation, invasion, and glycolysis of oral squamous cell carcinoma by sponged miR-424-5p. J Clin Lab Anal. 2022;36(7):e24486. doi: 10.1002/jcla.24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Tang K, Chen L, Du M, Qu Z. Exosomal CircGDI2 suppresses oral squamous cell carcinoma progression through the regulation of MiR-424-5p/SCAI axis. Cancer Manag Res. 2020;12:7501–7514. doi: 10.2147/CMAR.S255687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Wang G, Li Y, Zhao Q, Fan L, Tan B, et al. miR-424-5p reduces 5-fluorouracil resistance possibly by inhibiting Src/focal adhesion kinase signalling-mediated epithelial-mesenchymal transition in colon cancer cells. J Pharm Pharmacol. 2021;73(8):1062–1070. doi: 10.1093/jpp/rgab031. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Liang X, Xu J, Cai X. miR-424 targets AKT3 and PSAT1 and has a tumor-suppressive role in human colorectal cancer. Cancer Manag Res. 2018;10:6537–6547. doi: 10.2147/CMAR.S185789. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhang N, Li L, Luo J, Tan J, Hu W, Li Z, et al. Inhibiting microRNA-424 in bone marrow mesenchymal stem cells-derived exosomes suppresses tumor growth in colorectal cancer by upregulating TGFBR3. Arch Biochem Biophys. 2021;709:108965. doi: 10.1016/j.abb.2021.108965. [DOI] [PubMed] [Google Scholar]
- 34.Li N. CircTBL1XR1/miR-424 axis regulates Smad7 to promote the proliferation and metastasis of colorectal cancer. J Gastrointest Oncol. 2020;11(5):918–931. doi: 10.21037/jgo-20-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan W, Jiang X, Wang G, Li W, Zhai C, Chen S, et al. Cyto-biological effects of microRNA-424-5p on human colorectal cancer cells. Oncol Lett. 2020;20(4):120. doi: 10.3892/ol.2020.11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng C, Li H, Zheng J, Xu J, Gao P, Wang J. FENDRR sponges miR-424-5p to inhibit cell proliferation, migration and invasion in colorectal cancer. Technol Cancer Res Treat. 2020;19:1533033820980102. doi: 10.1177/1533033820980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shekhar R, Priyanka P, Kumar P, Ghosh T, Khan MM, Nagarajan P, et al. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J Biol Chem. 2019;294(12):4381–4400. doi: 10.1074/jbc.RA118.005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Gu J, Shen H, Shao T, Li S, Wang W, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA-424 in osteosarcoma. J Clin Lab Anal. 2020;34(2):e23045. doi: 10.1002/jcla.23045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long XH, Mao JH, Peng AF, Zhou Y, Huang SH, Liu ZL. Tumor suppressive microRNA-424 inhibits osteosarcoma cell migration and invasion via targeting fatty acid synthase. Exp Ther Med. 2013;5(4):1048–1052. doi: 10.3892/etm.2013.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Ruan Q, Zheng J, Zhang B, Yang H. LINC01116 promotes doxorubicin resistance in osteosarcoma by epigenetically silencing miR-424-5p and inducing epithelial-mesenchymal transition. Front Pharmacol. 2021;12:632206. doi: 10.3389/fphar.2021.632206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, Liu X. LINC00641 inhibits the development of cutaneous squamous cell carcinoma by downregulating miR-424 in A431 cells. Cancer Biother Radiopharm. 2021 doi: 10.1089/cbr.2020.4325. [DOI] [PubMed] [Google Scholar]
- 42.Han X, Jia Y, Chen X, Sun C, Sun J. lncRNA TINCR attenuates the proliferation and invasion, and enhances the apoptosis of cutaneous malignant melanoma cells by regulating the miR-424-5p/LATS1 axis. Oncol Rep. 2021;46(5):1–11. doi: 10.3892/or.2021.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Yamamoto Y, Takeshita F, Yamamoto T, Xiao Z, Ochiya T. Delivery of miR-424-5p via extracellular vesicles promotes the apoptosis of MDA-MB-231 TNBC cells in the tumor microenvironment. Int J Mol Sci. 2021;22(2):844. doi: 10.3390/ijms22020844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dastmalchi N, Safaralizadeh R, Hosseinpourfeizi MA, Baradaran B, Khojasteh SMB. MicroRNA-424-5p enhances chemosensitivity of breast cancer cells to Taxol and regulates cell cycle, apoptosis, and proliferation. Mol Biol Rep. 2021;48(2):1345–1357. doi: 10.1007/s11033-021-06193-4. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, Ji HW, Kim HW, Yun SH, Park JE, Kim SJ. Ginsenoside Rg3 prevents oncogenic long noncoding RNA ATXN8OS from inhibiting tumor-suppressive microRNA-424-5p in breast cancer cells. Biomolecules. 2021;11(1):118. doi: 10.3390/biom11010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie D, Song H, Wu T, Li D, Hua K, Xu H, et al. MicroRNA-424 serves an anti-oncogenic role by targeting cyclin-dependent kinase 1 in breast cancer cells. Oncol Rep. 2018;40(6):3416–3426. doi: 10.3892/or.2018.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, Yang T. Long non-coding RNA LINC00473 promotes breast cancer progression via miR-424-5p/CCNE1 pathway. Protein Pept Lett. 2022 doi: 10.2174/0929866530666221026164454. [DOI] [PubMed] [Google Scholar]
- 48.Dastmalchi N, Safaralizadeh R, Khojasteh SMB, Shadbad MA, Hosseinpourfeizi MA, Azarbarzin S, et al. The combined restoration of miR-424-5p and miR-142-3p effectively inhibits MCF-7 breast cancer cell line via modulating apoptosis, proliferation, colony formation, cell cycle and autophagy. Mol Biol Rep. 2022;49(9):8325–8335. doi: 10.1007/s11033-022-07646-0. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Wang S, Zhou J, Qian Q. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother. 2018;102:147–152. doi: 10.1016/j.biopha.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Chen B, Duan L, Yin G, Tan J, Jiang X. Simultaneously expressed miR-424 and miR-381 synergistically suppress the proliferation and survival of renal cancer cells–-Cdc2 activity is up-regulated by targeting WEE1. Clinics (Sao Paulo) 2013;68(6):825–833. doi: 10.6061/clinics/2013(06)17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D, Liu K, Li Z, Wang J, Wang X. miR-19a and miR-424 target TGFBR3 to promote epithelial-to-mesenchymal transition and migration of tongue squamous cell carcinoma cells. Cell Adh Migr. 2018;12(3):236–246. doi: 10.1080/19336918.2017.1365992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Liu H, Cui Y, Chen H, Cui X, Shao J, et al. miR-424-3p contributes to the malignant progression and chemoresistance of gastric cancer. Onco Targets Ther. 2020;13:12201–12211. doi: 10.2147/OTT.S280717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-β signaling pathway. Oncotarget. 2016;7(46):75185–75196. doi: 10.18632/oncotarget.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su J, Chen D, Ruan Y, Tian Y, Lv K, Zhou X, et al. LncRNA MBNL1-AS1 represses gastric cancer progression via the TGF-β pathway by modulating miR-424-5p/Smad7 axis. Bioengineered. 2022;13(3):6978–6995. doi: 10.1080/21655979.2022.2037921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui Y, Yang J, Bai Y, Zhang Y, Yao Y, Zheng T, et al. miR-424-5p regulates cell proliferation and migration of esophageal squamous cell carcinoma by targeting SIRT4. J Cancer. 2020;11(21):6337–6347. doi: 10.7150/jca.50587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F, Wang J, Yang X, Chen D, Wang L. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 2016;11(1):88. doi: 10.1186/s13000-016-0536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun YP, Lu F, Han XY, Ji M, Zhou Y, Zhang AM, et al. MiR-424 and miR-27a increase TRAIL sensitivity of acute myeloid leukemia by targeting PLAG1. Oncotarget. 2016;7(18):25276–25290. doi: 10.18632/oncotarget.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dallavalle C, Albino D, Civenni G, Merulla J, Ostano P, Mello-Grand M, et al. MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J Clin Invest. 2016;126(12):4585–4602. doi: 10.1172/JCI86505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu ZB, Shi SL, Pan FJ, Li L, Chen HY. Propranolol inhibits infantile hemangioma by regulating the miR-424/vascular endothelial growth factor-A (VEGFA) axis. Transl Pediatr. 2021;10(7):1867–1876. doi: 10.21037/tp-21-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, Li B, Feng J, Fang Q, Cheng J, Xie W, et al. JAG1, Regulated by microRNA-424-3p, involved in tumorigenesis and epithelial-mesenchymal transition of high proliferative potential-pituitary adenomas. Front Oncol. 2020;10:567021. doi: 10.3389/fonc.2020.567021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J, Zhao H, Zhang Q, Shi Z, Zhang Y, Zhao L, et al. Human papillomavirus type 16 E7 oncoprotein-induced upregulation of lysine-specific demethylase 5A promotes cervical cancer progression by regulating the microRNA-424-5p/suppressor of zeste 12 pathway. Exp Cell Res. 2020;396(1):112277. doi: 10.1016/j.yexcr.2020.112277. [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Jiao P, Zhong Y, Ji H, Zhang Y, Song H, et al. The endoplasmic reticulum-stressed head and neck squamous cell carcinoma cells induced exosomal miR-424-5p inhibits angiogenesis and migration of humanumbilical vein endothelial cells through LAMC1-mediated Wnt/β-catenin signaling pathway. Cell Transplant. 2022;31:9636897221083549. doi: 10.1177/09636897221083549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv HC, Lv YY, Wang G, Zhang XH, Li SN, Yue XF, et al. Mechanism of miR-424-5p promoter methylation in promoting epithelial-mesenchymal transition of hepatocellular carcinoma cells. Kaohsiung J Med Sci. 2022;38(4):336–346. doi: 10.1002/kjm2.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Z, Huang W, He J, Feng C. MicroRNA-424 inhibits autophagy and proliferation of hepatocellular carcinoma cells by targeting ATG14. Nan Fang Yi Ke Da Xue Xue Bao. 2021;41(7):1012–1021. doi: 10.12122/j.issn.1673-4254.2021.07.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu L, Yang F, Lin B, Chen X, Yin S, Zhang F, et al. MicroRNA-424 expression predicts tumor recurrence in patients with hepatocellular carcinoma following liver transplantation. Oncol Lett. 2018;15(6):9126–9132. doi: 10.3892/ol.2018.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Z, He J, Feng C. CircCBFB is a mediator of hepatocellular carcinoma cell autophagy and proliferation through miR-424-5p/ATG14 axis. Immunol Res. 2022;70(3):341–353. doi: 10.1007/s12026-021-09255-8. [DOI] [PubMed] [Google Scholar]
- 68.Shen X, Li Y, He F, Kong J. LncRNA CDKN2B-AS1 promotes cell viability, migration, and invasion of hepatocellular carcinoma via sponging miR-424-5p. Cancer Manag Res. 2020;12:6807–6819. doi: 10.2147/CMAR.S240000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C, et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1–11. doi: 10.1016/j.lfs.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 70.Ye Z, He Q, Wang Q, Lin Y, Cen K, Chen X. LINC00922 promotes the proliferation, migration, invasion and EMT process of liver cancer cells by regulating miR-424-5p/ARK5. Mol Cell Biochem. 2021;476(10):3757–3769. doi: 10.1007/s11010-021-04196-0. [DOI] [PubMed] [Google Scholar]
- 71.Wu L, Zhu X, Song Z, Chen D, Guo M, Liang J, et al. Long non-coding RNA HOXA-AS2 enhances the malignant biological behaviors in Glioma by epigenetically regulating RND3 expression. Onco Targets Ther. 2019;12:9407. doi: 10.2147/OTT.S225678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu M, Qin X, Zhou Y, Li G, Liu Z, Geng X, et al. Long non-coding RNA LINC00665 promotes gemcitabine resistance of Cholangiocarcinoma cells via regulating EMT and stemness properties through miR-424-5p/BCL9L axis. Cell Death Dis. 2021;12(1):72. doi: 10.1038/s41419-020-03346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu J, Yang B, Zhang Y, Feng X, He B, Xie H, et al. miR-424-5p represses the metastasis and invasion of intrahepatic cholangiocarcinoma by targeting ARK5. Int J Biol Sci. 2019;15(8):1591–1599. doi: 10.7150/ijbs.34113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu CT, Lin WY, Chang YH, Lin PY, Chen WC, Chen MF. DNMT1-dependent suppression of microRNA424 regulates tumor progression in human bladder cancer. Oncotarget. 2015;6(27):24119–24131. doi: 10.18632/oncotarget.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li WJ, Li G, Liu ZW, Chen ZY, Pu R. LncRNA LINC00355 promotes EMT and metastasis of bladder cancer cells through the miR-424-5p/HMGA2 axis. Neoplasma. 2021;68(6):1225–1235. doi: 10.4149/neo_2021_210427N574. [DOI] [PubMed] [Google Scholar]
- 76.Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. miR-34a, miR-424 and miR-513 inhibit MMSET expression to repress endometrial cancer cell invasion and sphere formation. Oncotarget. 2018;9(33):23253–23263. doi: 10.18632/oncotarget.25298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shu S, Liu X, Xu M, Gao X, Fan J, Liu H, et al. MicroRNA-424 regulates epithelial-mesenchymal transition of endometrial carcinoma by directly targeting insulin-like growth factor 1 receptor. J Cell Biochem. 2018 doi: 10.1002/jcb.27528. [DOI] [PubMed] [Google Scholar]
- 78.Wang P, Liu T, Zhao Z, Wang Z, Liu S, Yang X. SPTBN2 regulated by miR-424-5p promotes endometrial cancer progression via CLDN4/PI3K/AKT axis. Cell Death Discov. 2021;7(1):382. doi: 10.1038/s41420-021-00776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bieg D, Sypniewski D, Nowak E, Bednarek I. MiR-424-3p suppresses galectin-3 expression and sensitizes ovarian cancer cells to cisplatin. Arch Gynecol Obstet. 2019;299(4):1077–1087. doi: 10.1007/s00404-018-4999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho JG, Kim SW, Lee A, Jeong HN, Yun E, Choi J, et al. MicroRNA-dependent inhibition of WEE1 controls cancer stem-like characteristics and malignant behavior in ovarian cancer. Mol Ther Nucleic Acids. 2022;29:803–822. doi: 10.1016/j.omtn.2022.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li P, Xin H, Lu L. Extracellular vesicle-encapsulated microRNA-424 exerts inhibitory function in ovarian cancer by targeting MYB. J Transl Med. 2021;19(1):4. doi: 10.1186/s12967-020-02652-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu J, Gu Z, Tang Y, Hao J, Zhang C, Yang X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle. 2018;17(3):309–318. doi: 10.1080/15384101.2017.1407894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma LL, Liang L, Zhou D, Wang SW. Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma. 2021;68(1):165–173. doi: 10.4149/neo_2020_200707N705. [DOI] [PubMed] [Google Scholar]
- 84.Wu X, Ruan Y, Jiang H, Xu C. MicroRNA-424 inhibits cell migration, invasion, and epithelial mesenchymal transition by downregulating doublecortin-like kinase 1 in ovarian clear cell carcinoma. Int J Biochem Cell Biol. 2017;85:66–74. doi: 10.1016/j.biocel.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 85.Wu K, Hu G, He X, Zhou P, Li J, He B, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 86.Sun SL, Shu YG, Tao MY. LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol Cell Biochem. 2020;468(1–2):69–82. doi: 10.1007/s11010-020-03712-y. [DOI] [PubMed] [Google Scholar]
- 87.Wang L, Adiga A, Venkatramanan S, Chen J, Lewis B, Marathe M. Examining deep learning models with multiple data sources for COVID-19 forecasting. In: 2020 IEEE international conference on big data (big data), Atlanta, GA, USA. IEEE; 2020. p. 3846–3855.
- 88.Zhao C, Wang XB, Zhang YH, Zhou YM, Yin Q, Yao WC. MicroRNA-424 inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting KIF23 and functions as a novel prognostic predictor. Eur Rev Med Pharmacol Sci. 2018;22(19):6369–6378. doi: 10.26355/eurrev_201810_16049. [DOI] [PubMed] [Google Scholar]
- 89.Xu H, Zhang H, Tan L, Yang Y, Wang H, Zhao Q, et al. FAM87A as a competing endogenous RNA of miR-424-5p suppresses glioma progression by regulating PPM1H. Comput Math Methods Med. 2021;2021:7952922. doi: 10.1155/2021/7952922. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Zhao C, Zhao F, Chen H, Liu Y, Su J. MicroRNA-424-5p inhibits the proliferation, migration, and invasion of nasopharyngeal carcinoma cells by decreasing AKT3 expression. Braz J Med Biol Res. 2020;53(7):e9029. doi: 10.1590/1414-431X20209029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu C, Wang H, Chen S, Yang R, Li H, Zhang G. Baicalein inhibits cell growth and increases cisplatin sensitivity of A549 and H460 cells via miR-424-3p and targeting PTEN/PI3K/Akt pathway. J Cell Mol Med. 2018;22(4):2478–2487. doi: 10.1111/jcmm.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Lv Z, Fu J, Wang Z, Fan Z, Lei T. Endogenous microRNA-424 predicts clinical outcome and its inhibition acts as cancer suppressor in human non-small cell lung cancer. Biomed Pharmacother. 2017;89:208–214. doi: 10.1016/j.biopha.2017.01.163. [DOI] [PubMed] [Google Scholar]
- 93.Wen J, Hu Y, Liu Q, Ling Y, Zhang S, Luo K, et al. miR-424 coordinates multilayered regulation of cell cycle progression to promote esophageal squamous cell carcinoma cell proliferation. EBioMedicine. 2018;37:110–124. doi: 10.1016/j.ebiom.2018.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu X, Fu Y, Zhang G, Zhang D, Liang N, Li F, et al. miR-424-5p promotes anoikis resistance and lung metastasis by inactivating hippo signaling in thyroid cancer. Mol Ther Oncolytics. 2019;15:248–260. doi: 10.1016/j.omto.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–987. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 96.Hua F, Li CH, Chen XG, Liu XP. Long noncoding RNA CCAT2 knockdown suppresses tumorous progression by sponging miR-424 in epithelial ovarian cancer. Oncol Res. 2018;26(2):241–247. doi: 10.3727/096504017X14953948675412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bolandparva F, Hashemi Nasab MS, Mohamadnia A, Garajei A, Farhadi Nasab A, Bahrami N. Early diagnosis of oral squamous cell carcinoma (OSCC) by miR-138 and miR-424-5p expression as a cancer marker. Asian Pac J Cancer Prev. 2021;22(7):2185–2189. doi: 10.31557/APJCP.2021.22.7.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen B, Zhao Q, Guan L, Lv H, Bie L, Huang J, et al. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J Cell Mol Med. 2018;22(10):4751–4759. doi: 10.1111/jcmm.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long noncoding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR-424. Oncol Res. 2017;25(8):1391–1398. doi: 10.3727/096504017X14881559833562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teng F, Zhang JX, Chang QM, Wu XB, Tang WG, Wang JF, et al. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):235. doi: 10.1186/s13046-020-01739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao X, Zhang G, Li T, Zhou C, Bai L, Zhao J, et al. LINC00657 knockdown suppresses hepatocellular carcinoma progression by sponging miR-424 to regulate PD-L1 expression. Genes Genomics. 2020;42(11):1361–1368. doi: 10.1007/s13258-020-01001-y. [DOI] [PubMed] [Google Scholar]
- 102.Wang RP, Jiang J, Jiang T, Wang Y, Chen LX. Increased long noncoding RNA LINC00511 is correlated with poor prognosis and contributes to cell proliferation and metastasis by modulating miR-424 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(8):3291–3301. doi: 10.26355/eurrev_201904_17691. [DOI] [PubMed] [Google Scholar]
- 103.Yao J, Fu J, Liu Y, Qu W, Wang G, Yan Z. LncRNA CASC9 promotes proliferation, migration and inhibits apoptosis of hepatocellular carcinoma cells by down-regulating miR-424-5p. Ann Hepatol. 2021;23:100297. doi: 10.1016/j.aohep.2020.100297. [DOI] [PubMed] [Google Scholar]
- 104.Xu SJ, Xu WJ, Zeng Z, Zhang M, Zhang DY. MiR-424 functions as potential diagnostic and prognostic biomarker in melanoma. Clin Lab. 2020 doi: 10.7754/clin.lab.2019.190917. [DOI] [PubMed] [Google Scholar]
- 105.Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31(6):553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Z, Huang S, Lu W, Su Z, Yin X, Liang H, et al. Modeling the trend of coronavirus disease 2019 and restoration of operational capability of metropolitan medical service in China: a machine learning and mathematical model-based analysis. Glob Health Res Policy. 2020;5:20. doi: 10.1186/s41256-020-00145-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scholtz B, Horváth J, Tar I, Kiss C, Márton IJ. Salivary miR-31-5p, miR-345-3p, and miR-424-3p are reliable biomarkers in patients with oral squamous cell carcinoma. Pathogens. 2022;11(2):229. doi: 10.3390/pathogens11020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Richardsen E, Andersen S, Al-Saad S, Rakaee M, Nordby Y, Pedersen MI, et al. Low expression of miR-424-3p is highly correlated with clinical failure in prostate cancer. Sci Rep. 2019;9(1):10662. doi: 10.1038/s41598-019-47234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakashima T, Jinnin M, Etoh T, Fukushima S, Masuguchi S, Maruo K, et al. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS ONE. 2010;5(12):e14334. doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li MM, Dong CX, Sun B, Lei HZ, Wang YL, Gong YB, et al. LncRNA-MALAT1 promotes tumorogenesis of infantile hemangioma by competitively binding miR-424 to stimulate MEKK3/NF-κB pathway. Life Sci. 2019;239:116946. doi: 10.1016/j.lfs.2019.116946. [DOI] [PubMed] [Google Scholar]
- 111.Huang JJ, Zhuo F, Cai YH, Xiao H. Expression and clinical significance of serum MiR-424 and MiR-765 in multiple myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2022;30(2):461–465. doi: 10.19746/j.cnki.issn.1009-2137.2022.02.023. [DOI] [PubMed] [Google Scholar]
- 112.Yang C, Du P, Lu W. MiR-424 acts as a novel biomarker in the diagnosis of patients with hepatocellular carcinoma. Cancer Biother Radiopharm. 2021 doi: 10.1089/cbr.2020.4141. [DOI] [PubMed] [Google Scholar]
- 113.Lyu J, Zhao L, Wang F, Ji J, Cao Z, Xu H, et al. Discovery and validation of serum MicroRNAs as early diagnostic biomarkers for prostate cancer in Chinese population. Biomed Res Int. 2019;2019:9306803. doi: 10.1155/2019/9306803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiao CT, Lai WJ, Zhu WA, Wang H. MicroRNA derived from circulating exosomes as noninvasive biomarkers for diagnosing renal cell carcinoma. Onco Targets Ther. 2020;13:10765–10774. doi: 10.2147/OTT.S271606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Zhao L, Zhao P, Liu Z. Long non-coding RNA LINC00641 suppresses non-small-cell lung cancer by sponging miR-424-5p to upregulate PLSCR4. Cancer Biomark. 2019;26(1):79–91. doi: 10.3233/CBM-190142. [DOI] [PubMed] [Google Scholar]
- 116.Ding J, Zhang L, Chen S, Cao H, Xu C, Wang X. lncRNA CCAT2 enhanced resistance of glioma cells against chemodrugs by disturbing the normal function of miR-424. OncoTargets and Ther. 2020;13:1431–1445. doi: 10.2147/OTT.S227831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Han Y, Li X, Yan J, Ma C, Zheng X, Zhang J, et al. Knockdown of lncRNA PVT1 inhibits glioma progression by regulating miR-424 expression. Oncol Res Featur Preclin Clin Cancer Ther. 2019;27(6):681–690. doi: 10.3727/096504018X15424939990246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang Y-A, Huang Z-A, You Z-H, Zhu Z, Huang W-Z, Guo J-X, et al. Predicting lncRNA-miRNA interaction via graph convolution auto-encoder. Front Genet. 2019;10:758. doi: 10.3389/fgene.2019.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghonbalani ZN, Shahmohamadnejad S, Pasalar P, Khalili E. Hypermethylated miR-424 in colorectal cancer subsequently upregulates VEGF. J Gastrointest Cancer. 2022;53(2):380–386. doi: 10.1007/s12029-021-00614-0. [DOI] [PubMed] [Google Scholar]
- 120.Zhu B, Ju S, Chu H, Shen X, Zhang Y, Luo X, et al. The potential function of microRNAs as biomarkers and therapeutic targets in multiple myeloma. Oncol Lett. 2018;15(5):6094–6106. doi: 10.3892/ol.2018.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kanwal R, Plaga AR, Liu X, Shukla GC, Gupta S. MicroRNAs in prostate cancer: functional role as biomarkers. Cancer Lett. 2017;407:9–20. doi: 10.1016/j.canlet.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 122.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020;19(10):673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.