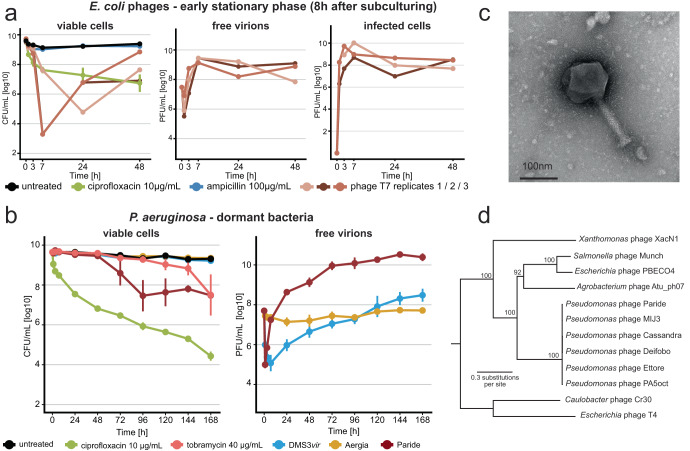
Fig. 2. Paride is a new P. aeruginosa phage that can replicate on deep-dormant host cells.
a E. coli K-12 MG1655 subcultured for 8 h were treated with either antibiotics or phage T7 (MOI ≈ 0.01) and viable colony forming units (CFU/ml) as well as plaque-forming units (PFU/ml) of free phages and infected cells were recorded over time. b P. aeruginosa PAO1 Δpel Δpsl subcultured for 48 h were treated with antibiotics or phages (MOI ≈ 0.01) and viable CFU/ml as well as free phages were recorded over time (see also Supplementary Note 1). All data points and error bars show the average of three biological replicates and standard error of the mean except in (a), where qualitatively similar but temporally shifted results of T7 infection experiments are shown individually. Limits of detection are 2 log10 CFU/mL for viable cells and 3.6 log10 PFU/mL for free phages. c Negative stain TEM micrograph of phage Paride. d Maximum-likelihood phylogeny of Paride and other group 2.2 jumbo phages as defined by Iyer et al.42 with phages T4 and Cr30 as outgroup (see “Methods”). Bootstrap support is shown if >70. Source data are provided as a Source data file.