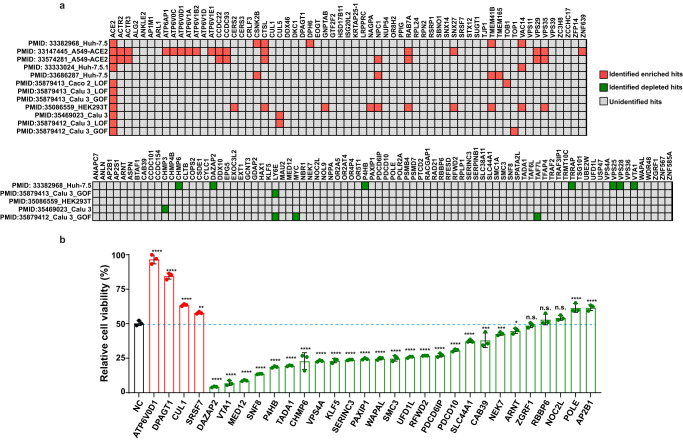
Fig. 3. Validation of host factors identified from the CRISPR dropout screen.
a Performance of identified host factors in previously reported SARS-CoV-2 screens. For each dataset, enriched and depleted hits that meet the listed criteria were marked as red and green squares, respectively. Others that fail to be identified in the listed datasets were marked as grey squares. b Effects of perturbation of top 30 hits on the CPE caused by SARS-CoV-2 infection. Four pro-viral and 26 anti-viral factors were selected for validation. A set of A549-AC cell lines expressing gene-specific gRNAs were infected with recombinant SARS-CoV-2 at MOI = 2.5. The percentages of viable cells were measured at 48 h post-infection. Data were normalized using the viability of corresponding cells at mock conditions. Statistical significance between cells expressing gene-specific gRNAs and non-targeting gRNA (NC) was determined by one-way ANOVA with repeated measurements. At least two independent experiments were performed, and samples were triplicated in each independent experiment. Data were analyzed using one-way ANOVA followed by Dunnett’s post hoc test and were presented as mean values ± SD; n = 3 biologically independent samples. Exact P values were provided in the source data file. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. n.s. not significant.