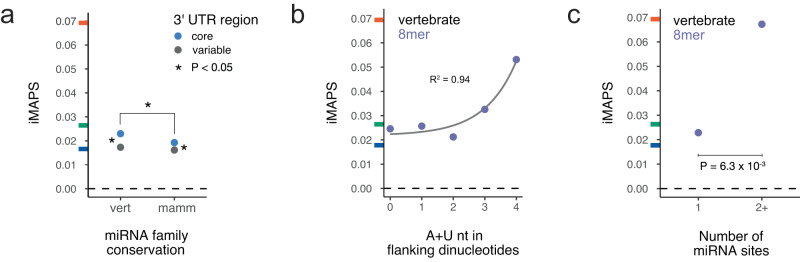
Fig. 4. Negative selection at miRNA target sites in 3ʹ UTRs.
a miRNA targets (including 8mer, 7mer-m8, and 7mer-A1 target types) are under selection in core 3ʹ UTR regions for both broadly conserved (“vert” = vertebrate) and conserved (“mamm” = mammalian) microRNA families. n = 179,220 variants for vertebrate core, 197,573 for mammalian core, 137,103 for vertebrate variable, and 147,679 for mammalian variable. Asterisks (*) indicate one-sided Fisher Exact Tests with P < 0.05. Vertebrate P = 4.3 ×10−15. Mammalian P = 1.6 ×10−5. b Stronger selection is observed for 8mer targets with increasing AU content of the dinucleotides immediately preceding and following the target site. The gray line is a linear model describing the data using an exponential fit. R2 shows Pearson’s product-moment correlation. c Variants within targets recognized by two or more broadly conserved miRNA families are under stronger selection than those within targets of a single miRNA family. n = 10,912 variants for single targets and n = 389 variants for variants in targets of multiple miRNAs. A one-sided Fisher Exact Test P value is shown. Tick marks on the y-axis indicate the genome-wide average iMAPS for 3ʹ UTR (blue), synonymous (green), and missense (orange) coding variants. The dashed line at 0 indicates background levels of intergenic selection.