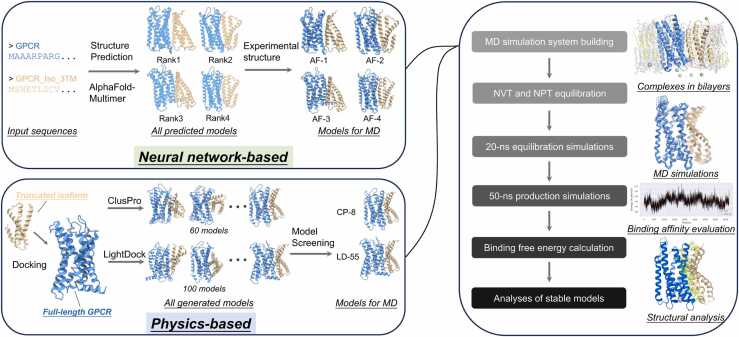
Fig. 1.
The workflow of our computational approach. Briefly, we generated initial complex models by combining neural network-based protein structure prediction tool AlphaFold2 and two physics-based protein-protein docking tools, ClusPro and LightDock. For complex structure prediction, we employed AlphaFold-Multimer to generate five models, out of which the models with unique interface composition were selected. These models were then aligned with experimental structures to replace predicted structures (except isoforms). For protein-protein docking, ClusPro generated 60 models and LightDock generated 100 models, from which topology-compliant models were chosen. Both sets of initial complex models were used as inputs to build protein complex-membrane systems for molecular dynamics (MD) simulations. After NVT and NPT equilibration, a 20-ns equilibration MD simulation was run to achieve stabilization of the complexes. This was followed by a 50-ns production MD simulation, subsequently subjected to binding free energy calculations based on MMPBSA algorithm. According to the results, the most stable complex models were selected for further analysis. The structure snapshots were made using PyMOL or ChimeraX.