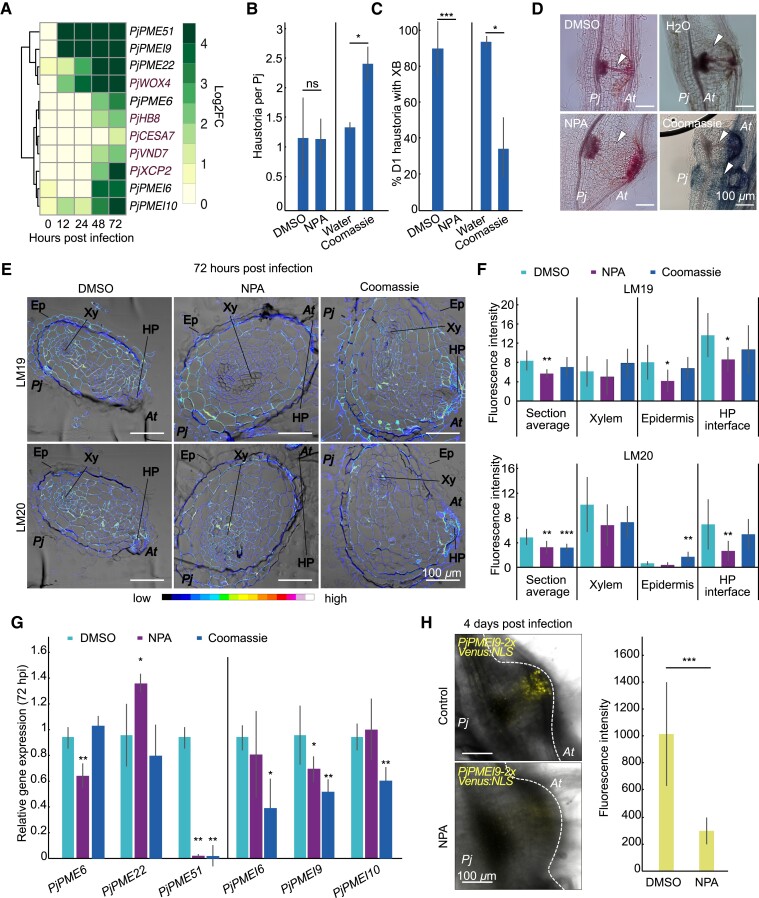
Figure 6.
PjPMEs and PjPMEIs expression associates with xylem bridge development. A) Expression heatmap of P. japonicum cambium and xylem marker genes (red) and selected PjPMEs and PjPMEIs: log2 fold change between P. japonicum infecting and not infecting over five time points during infection, clustered by expression. B) Number of haustoria per P. japonicum plant at 7 dpi during treatment with DMSO, 5 μM NPA (n = 2 replicates), water or 0.05 mM Coomassie (n = 3 replicates). C) Percentage of day one (D1) haustoria with a xylem bridge (XB) formed during treatment with DMSO, 5 μM NPA (n = 2 replicates), water or 0.05 mM Coomassie (n = 3 replicates). D) Images of 7 dpi haustoria formed on DMSO, 5 μM NPA, water or 0.05 mM Coomassie. White arrowheads denote the location of XB development. E) Fluorescence images of antibody staining using LM19 and LM20 on cross sections of 72 hpi haustoria developed on DMSO, 5 μM NPA or 0.05 mM Coomassie + DMSO. Ep = epidermis, Xy = xylem, HP = host-parasite interface. At = A. thaliana, Pj = P. japonicum. F) Fluorescence quantification in whole P. japonicum sections (section average), host-parasite (HP) interface, xylem (root xylem and xylem bridge) and epidermis tissues for LM19 and LM20 antibodies. Asterisks indicate significance compared to DMSO (Student's t-test, P-value corrected for multiple testing, n = 8 to 14 sections). G) Relative gene expression of select PjPMEs and PjPMEIs at 72 hpi in P. japonicum haustoria treated with 5 μM NPA or 0.05 mM Coomassie + DMSO normalised to DMSO (n = 3 replicates). H) Images of haustoria developed on hairy roots expressing the PjPMEI9 nuclear-localised (NLS) transcriptional reporter at 4 dpi on DMSO or 5 μM NPA, and quantification of fluorescence intensity (n = 7 to 10 haustoria). Scale bars 100 μm. For all panels, asterisks indicate significance compared to control (Student's t-test): * for P < 0.05, ** for P < 0.01, *** for P < 0.001, bars represent standard deviation.