Abstract
Expansins facilitate cell expansion by mediating pH-dependent cell wall (CW) loosening. However, the role of expansins in controlling CW biomechanical properties in specific tissues and organs remains elusive. We monitored hormonal responsiveness and spatial specificity of expression and localization of expansins predicted to be the direct targets of cytokinin signaling in Arabidopsis (Arabidopsis thaliana). We found EXPANSIN1 (EXPA1) homogenously distributed throughout the CW of columella/lateral root cap, while EXPA10 and EXPA14 localized predominantly at 3-cell boundaries in the epidermis/cortex in various root zones. EXPA15 revealed cell-type-specific combination of homogenous vs. 3-cell boundaries localization. By comparing Brillouin frequency shift and AFM-measured Young's modulus, we demonstrated Brillouin light scattering (BLS) as a tool suitable for non-invasive in vivo quantitative assessment of CW viscoelasticity. Using both BLS and AFM, we showed that EXPA1 overexpression upregulated CW stiffness in the root transition zone (TZ). The dexamethasone-controlled EXPA1 overexpression induced fast changes in the transcription of numerous CW-associated genes, including several EXPAs and XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASEs (XTHs), and associated with rapid pectin methylesterification determined by in situ Fourier-transform infrared spectroscopy in the root TZ. The EXPA1-induced CW remodeling is associated with the shortening of the root apical meristem, leading to root growth arrest. Based on our results, we propose that expansins control root growth by a delicate orchestration of CW biomechanical properties, possibly regulating both CW loosening and CW remodeling.
Expansins control root growth by orchestrating cell wall biomechanical properties, possibly regulating cell wall stiffening and root growth arrest associated with cell wall remodeling.
Introduction
The cell wall (CW) is a fundamental component of plant cells that shapes the plant body and plays key roles in the development and growth of organs, the movement of solutes and nutrients, and in protecting plants from the environment. The CW is also important in controlling cell differentiation and intercellular communication during development (Wolf et al. 2012). CWs provide rigidity; however, they also have the ability to expand. Recent studies of growth regulatory networks suggest that turgor-driven cell expansion is the result of a fine-tuned balance between wall relaxation and stiffening linked via a mechanosensing feedback loop (Sassi and Trass 2015; Braybrook and Jönsson 2016; Willis et al. 2016; Trinh et al. 2021). These regulatory networks comprise transcription factors and plant hormones and allow tight control over the equilibrium between cell division and differentiation, a process fundamental to the growth and development of individual organs in any multicellular organism.
The primary CW consists predominantly of polysaccharides—cellulose, hemicellulose, and pectins. While cellulose microfibrils are responsible for the main load-bearing characteristics of the CW, the presence of hemicellulose and pectins can alter viscoelastic properties of the wall matrix (Wolf et al. 2012; Cosgrove 2018a; Zhang et al. 2021). Once the final cell size is reached, a secondary CW can be deposited in specific cell types, e.g. xylem tracheary elements (for review, see Didi et al. 2015), to provide additional mechanical resistance. CW mechanical properties are modulated by controlling the biochemical composition, as well as the degree and nature of linkages between the CW polysaccharides. Interestingly, wall extensibility may be controlled in discrete regions—so-called biomechanical hotspots—where cellulose–cellulose contacts are made, potentially mediated by trace amounts of xyloglucan (Cosgrove 2014, 2018a, 2018b). These relatively few contact points between cellulose microfibrils may be key sites of CW loosening—a complex process allowing targeted wall expansion.
Expansins, originally described as CW loosening agents during “acid growth” (McQueen-Mason et al. 1992), become activated during CW acidification triggered by a number of stimuli acting on the plasma membrane H+-ATPase proton pump (Cosgrove 2005). Mechanistically, expansin-mediated CW loosening neither requires polysaccharide hydrolytic activity nor changes in the composition of the CW; instead, expansins are proposed to disrupt non-covalent bonds between cellulose and components of the surrounding CW matrix, most probably xyloglucans, thus relaxing wall stresses and allowing turgor-driven expansion (McQueen-Mason et al. 1995; Cosgrove 2005; Park and Cosgrove 2012). Arabidopsis (Arabidopsis thaliana) has 36 members of the expansin superfamily, 26 of them belonging to the family of α-expansins (EXPA) that were experimentally proven to promote CW loosening (Li et al. 2002; Sampedro and Cosgrove 2005). EXPA1 (At1g69530) has been known for decades from experiments with beads loaded with purified expansin that induced leaf primordia formation on the tomato shoot apical meristem (Fleming et al. 1997). Apart from leaf organogenesis (Reinhardt et al. 1998) and vascular tissue differentiation (Cho and Kende 1998), expansins are involved in root development and growth (Lee and Kim 2013; Pacifici et al. 2018; Ramakrishna et al. 2019), root hair initiation (Cho and Cosgrove 2002), and seed germination (Ribas et al. 2019; Sanchez-Montesino et al. 2019); for more examples, see the recent review by Samalova et al. (2022). Interestingly, NbEXPA1 was shown to be plasmodesmata-specific and functions as a host factor for potyviral infection (Park et al. 2017).
Biomechanical interactions of cells with the extracellular matrix have been shown to be an important regulator of cell fate specification in animal models (Engler et al. 2006; Yang et al. 2016). In plants, changes in CW mechanics are the driving force of growth and development, as predicted by a number of biomechanical models (Geitmann and Ortega 2009; Hamant and Traas 2010; Sassi and Traas 2015; Braybrook and Jonsson 2016; Haas et al. 2020). For instance, in vivo chemical modification (demethylesterification) of homogalacturonan by pectin methylesterases was shown to be sufficient for the initiation of novel flower and floral organ primordia in Arabidopsis. In contrast, inhibition of homogalacturonan demethylesterification resulted in the inhibition of normal organ formation (Peaucelle et al. 2008). Importantly, demethylesterification of homogalacturonan was shown to be associated with an increase in CW plasticity as measured via atomic force microscopy (AFM), suggesting that higher elasticity of CWs might be instructive for newly formed organ primordia (Peaucelle et al. 2011). Nonetheless, the opposite response, i.e. CW stiffening, has also been observed upon pectin demethylesterification (reviewed in Levesque-Tremblay et al. 2015). In plants, the importance of biomechanical CW properties has been described mostly in shoot tissues (Reinhardt et al. 1998; Pien et al. 2001; Hamant et al. 2008; Sampathkumar et al. 2014; Landrein et al. 2015; Gruel et al. 2016; Hervieux et al. 2017; Majda et al. 2017; Takatani et al. 2020). However, the biomechanical interactions associated with controlling CW properties are emerging as an important mechanism also guiding root growth and development (Vermeer et al. 2014; Barbez et al. 2017; Pacifici et al. 2018; Ramakrishna et al. 2019; Hurny et al. 2020).
Phytohormones, including auxins and cytokinins, are key players in growth regulation responses and are thus determinants of plant architecture and CW development. The role of auxins in the “acid growth theory” is well known (Cleland 1971; Hager et al. 1971), in which auxin-induced extrusion of protons into the apoplast activates expansins, leading to CW loosening and growth. Recently, auxin signaling has been shown to play a role in both transcriptional and non-transcriptional regulations leading to apoplast acidification (reviewed in Li et al. 2021). Cytokinins are factors controlling the equilibrium between cell division and differentiation (Dello Ioio et al. 2007, 2008) in the root apical meristem (RAM) by delimiting the auxin minimum that triggers the developmental switch (Di Mambro et al. 2017). Pacifici et al. (2018) proposed that EXPA1 is a direct target of multistep phosphorelay signaling in cytokinin-regulated cell differentiation in the RAM. Recently, cytokinins were also implicated in root growth control, attenuating cell elongation by inducing CW stiffening (Liu et al. 2022).
Brillouin light scattering (BLS) microscopy is an all-optical label-free spectroscopic technique that allows one to spatially map the inelastic Brillouin frequency shift (BFS, νΒ) with a lateral resolution near the diffraction limit (Elsayad et al. 2019; Prevedel et al. 2019), and has been used on live cells (Scarcelli et al. 2015) and tissues (Elsayad et al. 2016, Bacete et al. 2022). The distance of the Brillouin peaks (in GHz) from the central laser frequency is a measure of the local mechanical properties within the confocal volume. Despite probing a distinct elastic modulus in a different frequency regime, the measured BLS has been empirically found to (semi-logarithmically) scale with the lower frequency stiffness defined by Young's modulus. BFS can be expected to be higher for “stiffer” samples and smaller for “softer” cells and tissue (Scarcelli et al. 2015; Andriotis et al. 2019; Gouveia et al. 2019). Young's modulus is typically measured by AFM. This method is based on micrometer or nanometer tissue compressions (indentations) and was developed to precisely measure the mechanical properties of CWs in developing organs and across entire tissue regions (Peaucelle 2014). The measured stiffness (resistance to deformation/indentation) is defined by the measurement of an indentation modulus that best describes the elasticity of the CW scaffolding in the given tissue. AFM can be also used to image CW surface topology at high resolution to detect individual cellulose microfibrils (app. 3 nm in diameter, Zhang et al. 2014) and can be carried out under water, allowing CW imaging of under near-native conditions.
In this work, we set out to localize EXPA1 and its homologs EXPA10, EXPA14, and EXPA15 in planta and describe the relationship between the expression of expansins and CW mechanical properties during root cell differentiation. Using both BLS and the more established AFM, we demonstrate CW stiffening upon EXPA1 overexpression accompanied by changes in CW chemical composition leading to remarkable root shortening in Arabidopsis seedlings.
Results
Cytokinins and auxins control EXPA transcription in the Arabidopsis root
For our study, we selected the following expansins, AtEXPA1 (EXPA1, At1g69530), AtEXPA10 (EXPA10, At1g26770), AtEXPA14 (EXPA14, At1g69530), and AtEXPA15 (EXPA15, At2g03090), that have previously been suggested to be under hormonal control (Lee et al. 2007; Bhargava et al. 2013; Pacifici et al. 2018; Ramakrishna et al. 2019).
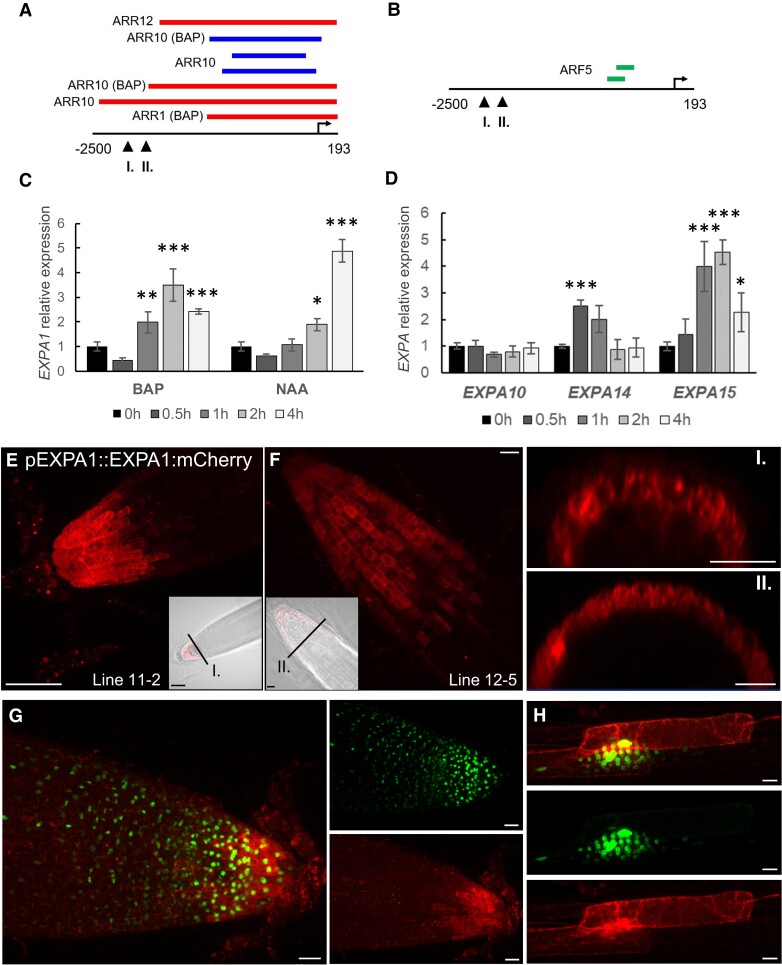
Type-B Response Regulator binding assays and cytokinin responsiveness (Taniguchi et al. 2007; Zubo et al. 2017; Pacifici et al. 2018; Xie et al. 2018) indicate that EXPA1 is the direct target of cytokinin-responsive ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) and its homologs ARR10 and ARR12 (Fig. 1A). EXPA1 responsiveness to auxin could be potentially regulated by AUXIN RESPONSE FACTOR 5 (ARF5) since the corresponding DNA affinity purification (DAP)-sequencing peaks (O’Malley et al. 2016) are located in its promoter (Fig. 1B). To confirm hormonal control over EXPA1, we quantified EXPA1 transcripts using reverse transcription quantitative PCR (RT-qPCR) in wild-type (WT) Arabidopsis seedlings treated with 5 μM 6-benzylaminopurine (BAP) or 5 μM 1-naphthaleneacetic acid (NAA, Fig. 1C, Supplemental Materials and Methods, Supplemental Table S1). With the cytokinin treatment, transcript levels were transiently and moderately (3 to 4 times compared with mock-treated control) upregulated over the 4-h time span tested. By comparison, EXPA1 responded slightly more distinctly to the auxin treatment, and its transcript level increased continuously up to 5- to 10-fold at 4 h. However, no or only very weak increase in EXPA1 promoter activity was detected after cytokinin or auxin treatment, respectively (Supplemental Fig. S1A), in either the RAM or the transition zone (TZ) in a pEXPA1::nls:3xGFP transcriptional fusion line (Ramakrishna et al. 2019).
Figure 1.
EXPA1 promoter activity and its localization in roots, and transcript profiling of expansins in response to hormones. A)EXPA1 promoter analysis identifies ChIP-seq- and DAP-seq-derived binding events for transcription factors involved in cytokinin and (B) auxin signaling pathways. In both (A) and (B) red, blue, and green colors depict peaks from Xie et al. (2018), Zubo et al. (2017), and O’Malley et al. (2016), respectively. The coordinates represent positions relative to the transcription start site marked by the arrow. Arrowheads indicate the 5′-end of the pEXPA1 promoter in this publication (I.) and Pacifici et al. (2018) (II.). C) Quantitative real-time PCR and EXPA1 transcript abundance in 7-d-old Arabidopsis WT roots treated with 5 μM BAP or 5 μM NAA for 0.5, 1, 2, and 4 h. D) Quantitative real-time PCR-assayed EXPA10, EXPA14, and EXPA15 transcript abundance in 7-d-old Arabidopsis WT roots treated with 5 μM BAP for 0.5, 1, 2, and 4 h. In both (C) and (D), EXPAs transcript abundance is double normalized to UBQ10 and mock-treated controls. The experiment was repeated twice with 3 replicas of each sample (n = 6), and error bars represent SD. Both experiments were evaluated using simple ANOVA and post hoc Tukey test. Statistically significant differences at alpha 0.05 (*), 0.01 (**), and 0.001 (***) are shown. E, F) Z-stack projections of pEXPA1::EXPA1:mCherry fusion localized in the LRC and columella of 2 independent single-copy transgenic lines 11-2 (D) and 12-5 (E) and their transversal xz optical sections (I. and II.) as indicated by the black lines in the transmitted-light micrograph inserts shown as a single optical section. G, H) Z-stack projections of F1 line pEXPA1::EXPA1:mCherry (11-2) crossed with pEXPA1::nls:3xGFP illustrating a similar pattern of EXPA1 expression by GFP (green) and mCherry (red) signals in the RAM (F) and the lateral root primordium (G). Scale bars correspond to 20 μm except in (D) and inset I., where it corresponds to 50 μm.
EXPA10, EXPA14, and EXPA15 were upregulated by exogenous cytokinins in the WT Col-0, and this induction was reduced in the line overexpressing ARR7, type-A ARR, and the negative regulator of cytokinin signaling (Lee et al. 2007). Based on our in silico analysis, EXPA10, EXPA14, and EXPA15 might also be direct targets of cytokinin-activated type-B ARRs (Supplemental Fig. S1B). In line with this finding, both EXPA14 and EXPA15 were upregulated by cytokinins; nonetheless, in contrast to a previous report (Pacifici et al. 2018), no positive response was detected with EXPA10 (Fig. 1D).
Altogether, our data suggest moderate and transient EXPA1 and EXPA14 upregulation by cytokinins (in the case of EXPA1 taking place outside the RAM/TZ), with a stronger response being seen for auxin- and cytokinin-mediated upregulation of EXPA1 and EXPA15, respectively. However, no clear effect of exogenous hormone application was detectable for EXPA10.
Expansins localize to the root CW in a specific pattern
Expansins were previously shown to be localized in the CW by immunogold labeling of CWs and Golgi-derived vesicles using an antibody against α-expansin (Cosgrove et al. 2002). However, attempts to visualize expansins in the CW of living plants using a translational fusion with a green fluorescent protein (GFP) have so far failed (Pacifici et al. 2018), perhaps due to the high sensitivity of GFP to the low pH environment. Therefore, we created translational fusions of EXPA1, EXPA10, EXPA14, and EXPA15 with the red fluorescent protein mCherry (Shaner et al. 2004; Supplemental Table S2) under the control of native promoters and confirmed their CW localization in a highly tissue-specific manner in roots (Figs. 1 and 2).
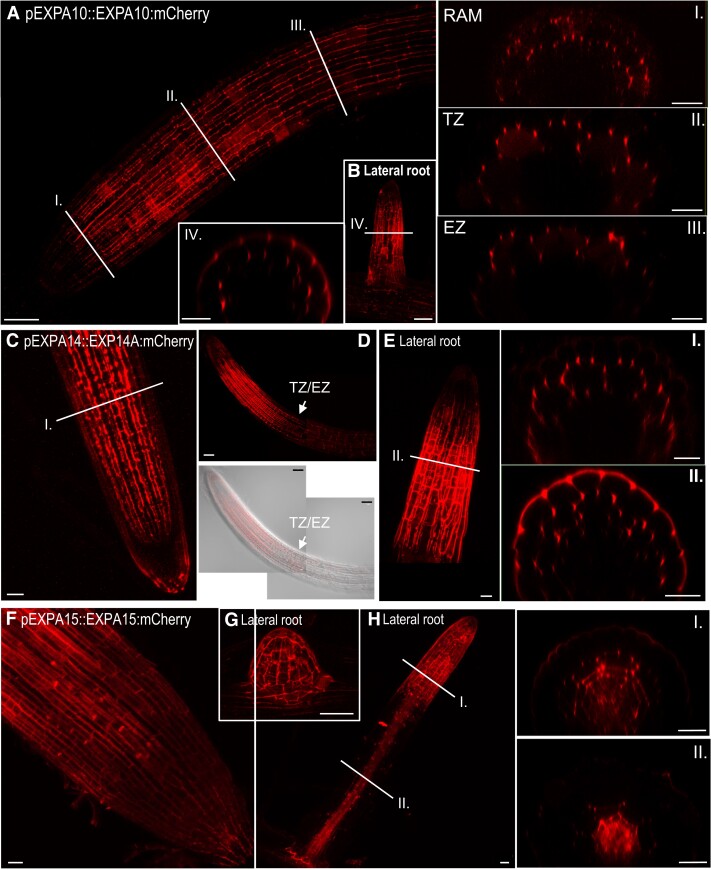
Figure 2.
EXPA10, EXPA14, and EXPA15 reveal specific localization in the root tip. A, B)pEXPA10::EXPA10:mCherry in the primary (A) and lateral (B) root and transversal xz optical sections (I.-IV.) through the RAM, transition (TZ) and elongation (EZ) zones, and the lateral root as indicated by white lines. C–E)pEXPA14::EXPA14:mCherry in the primary (C, D) and lateral (E) root and transversal xz optical sections (I. and II.) as indicated by white lines; white arrows point to the TZ/EZ boundary. The specimen in the bottom panel of (D) is shown by composite images comprised of 2 partially overlapping fields of view. F–H)pEXPA15::EXPA15:mCherry in RAM (F), emerging lateral root (G), lateral root (H), and its transversal xz optical sections (I. and II.) as indicated by white lines. Z-stack projections from optical sections taken from the top of the root to the center are shown in all longitudinal plane views. Transmitted-light micrograph inserts show a single optical section. Scale bars correspond to 20 μm except in (A, B, D, G), where they correspond to 50 μm.
In Arabidopsis root, EXPA1 revealed the strongest expression in the columella and lateral root cap (LRC) of both the primary and lateral roots (Fig. 1, E to G). Interestingly, cells immediately surrounding the developing lateral roots and primordia also strongly expressed EXPA1 (Fig. 1H). These results were confirmed using an independent transcriptional pEXPA1::nls:3xGFP fusion line (Ramakrishna et al. 2019) crossed into the mCherry line background (Fig. 1, G and H). Occasionally, very weak EXPA1 promoter activity was detectable in the boundary between the root TZ and the elongation zone (EZ) (Supplemental Fig. S2A); however, no EXPA1:mCherry was detectable there (Supplemental Fig. S2B).
EXPA10 was visibly expressed in the cortex layer of the primary root from the meristematic zone up (proximally) to the first lateral roots (Fig. 2A). Unlike EXPA1, EXPA10 was not expressed in the LRC. Interestingly, in contrast to the rather homogenous distribution of EXPA1 throughout the CW surrounding the LRC/columella cells, we observed a distinct “spotty” localization of EXPA10, predominantly in the cortex/endodermis and cortex/epidermis 3-cell boundaries that were particularly visible on cross-sections of the roots (Fig. 2A, insets I. to III.). In the lateral roots, the EXPA10:mCherry fusion localized predominantly to the epidermis and cortex layers of the transition/elongation zones (Fig. 2B and inset IV.).
EXPA14:mCherry fusion localized to the cortex layer of the RAM up to the TZ/EZ boundary, with the signal not being detectable more proximally (Figs. 2, C and D). The distinct pattern of strong accumulation of the protein in the apoplastic space at the boundary of 3 cells (insets I. and II. for Fig. 2, C and E, Supplemental Fig. S3A) resembles the one observed for the EXPA10:mCherry fusion. In lateral roots, EXPA14 is expressed dominantly in the TZ/EZ boundary (Fig. 2E). However, in contrast to the primary root, EXPA14 is located not only in the lateral root cortex, but also in the epidermal cell layers (Fig. 2E, inset II.).
EXPA15:mCherry was localized to the RAM epidermis (Fig. 2F) and emerging lateral roots (Fig. 2G, Supplemental Fig. S3B) in a relatively uniform pattern; however, the “spotty” pattern at the 3-cell boundary was apparent in the internal cortex/endodermis. Proximally from the meristematic zone, EXPA15 re-localized into deeper (vasculature) layers, again showing a rather homogenous distribution (Fig. 2H, insets I. to II.). It is worth mentioning that in contrast to homogenously distributed EXPA1 and (partially) EXPA15, EXPA10 and EXPA14 seem to be localized mainly in the CWs oriented in parallel with the longitudinal root axis walls in the RAM/TZ of the main root (Supplemental Fig. S3C).
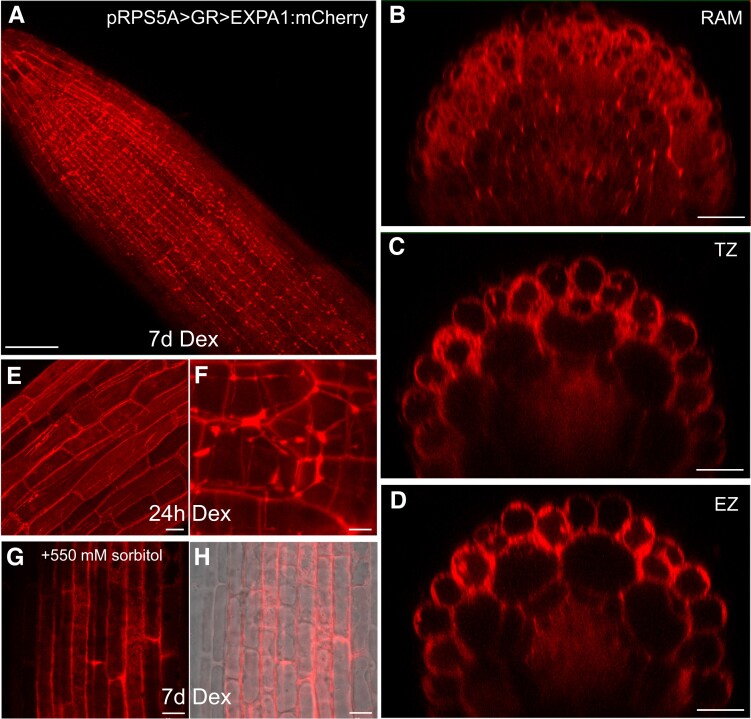
To confirm the extracellular localization, we activated the (naturally very weak) expression of EXPA1:Cherry in all plant tissues (Fig. 3) using the dexamethasone (Dex) inducible pOp6/LhGR system (Craft et al. 2005; Samalova et al. 2005, et al2019). After both long (7 d) and short (24 h) Dex induction, the fusion protein accumulated in the cell periphery/apoplastic space in roots but was also visible in the transit through the secretory pathway from the endoplasmic reticulum to the CW. However, because the resolution of a confocal microscope did not allow us to distinguish between CW and plasma membrane localization, we treated the roots with 550 mM sorbitol to allow for plasmolysis. Figure 3G shows that unlike the plasma membrane marker (Supplemental Fig. S4), EXP A1:mCherry remained located at the outer edges of the cells, suggesting that EXPA1 is indeed localized in the CW. Importantly, the CW localization pattern we observed in the case of the Dex-induced pRPS5A > GR > EXPA1:mCherry line still resembled the homogenous distribution of EXPA1:mCherry driven by its natural promoter in the LRC (compare Figs. 1 and 3). This suggests that the CW localization pattern of EXPA1 is independent of cell type and level of expression.
Figure 3.
Overproduced EXPA1 localizes to the CW. Z-stack projections of EXPA1:mCherry fluorescence in pRPS5A>GR>EXPA1:mCherry seedlings induced by Dex on solid MS medium for 7d in the primary root (A) and its xz optical cross-sections through RAM (B), TZ (C), and EZ (D). E, F)pRPS5A>GR>EXPA1:mCherry seedlings induced by Dex in liquid MS medium for 24 h; EZ of the primary root (E) and lateral root (F). G, H)pRPS5A>GR>EXPA1:mCherry seedlings induced as in (A) and after 10 min-treatment of the primary root with 550 mM sorbitol. Fluorescence channel (G) and its overlay with transmitted light (H). Scale bars correspond to 20 μm except in (A) (50 μm), (F) (5 μm), and (G & H) (10 μm).
To conclude, all assayed expansins show distinct expression and localization patterns. The differences in the localization pattern between EXPA1 revealing homogenous distribution all around the cell and the “spotty” localization of EXPA10, EXPA14, and (partially) EXPA15 suggest differential roles of individual EXPAs in controlling root CW properties.
EXPA1 overexpression stiffens the root CW
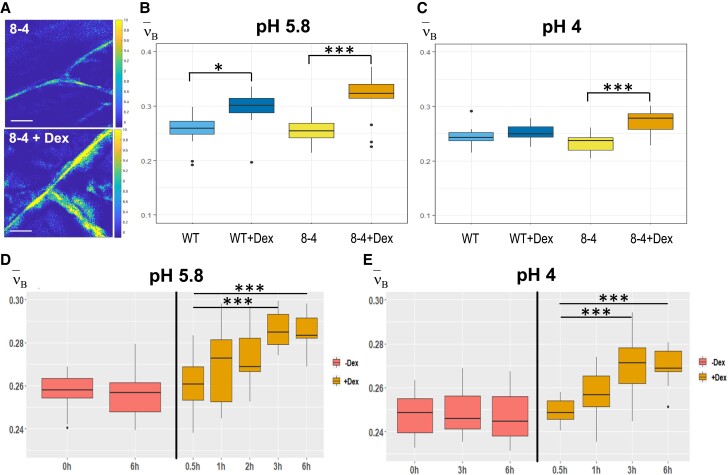
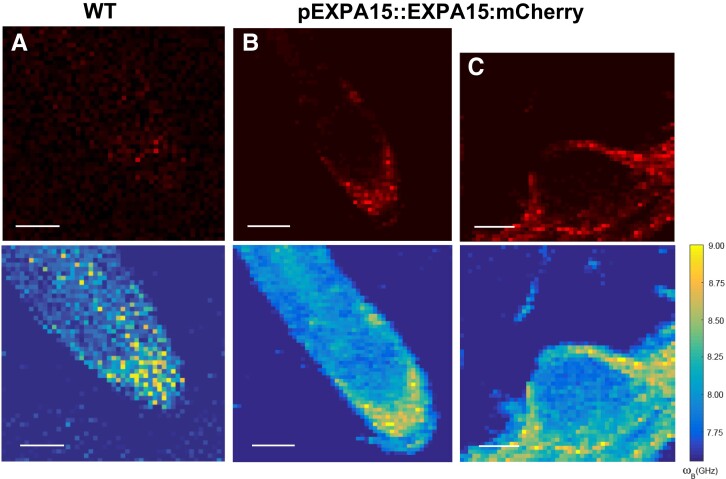
To characterize its potential importance in controlling CW biomechanical properties, we overexpressed EXPA1 (without any tag) and generated pRPS5A>GR>EXPA1 Dex-inducible lines (8-4 and 5-4) using the pOp6/LhGR system as above. Representative 2D BFS maps (Fig. 4A) display the BLS shift in the CW of plants overexpressing EXPA1 before and after induction. We quantified the ν'Β in roots (TZ/EZ boundary) of 7-d-old Arabidopsis WT and EXPA1 overexpressing seedlings (line 8-4) grown on MS media pH 5.8 or pH 4 (Fig. 4, B and C, respectively) with or without Dex induction. For technical reasons (to obtain sufficient overlap of point spread function with the CW and, hence, a good CW signal), and following the expression profile of assayed EXPA genes (epidermis and/or cortex), we focused on the anticlinal portion of longitudinal CWs in the root epidermis. The plants overexpressing EXPA1 showed a higher Brillouin Elastic Contrast [ν'Β, a BFS normalized to the BFS of distilled water, a measure of longitudinal elastic modulus (Antonacci et al. 2020); see Materials and Methods], for the root CWs on both pH media, suggesting that their CWs are stiffer. Dex-induced stiffening was also observed in the WT at pH 5.8. However, this effect was smaller compared with the ν'Β in line 8-4 and was not observed under pH 4 (Fig. 4, B and C). The pRPS5A>GR>EXPA1:mCherry lines induced on Dex also displayed higher ν'Β, but it was not significantly different from non-induced plants (Supplemental Fig. S5A), perhaps due to lower expression levels of the EXPA1:mCherry (Supplemental Fig. S5B). It may also be due to the possibility that the recombinant protein is less functional in plants. Also, these data clearly demonstrate no effect of Dex alone on the CW stiffening. Importantly, we detected an increase in the ν'Β (CW stiffness) in response to even short-term Dex induction (up to 6 h) that was sufficient to upregulate EXPA1 expression (Supplemental Fig. S6A) and EXPA1:mCherry fluorescence (Supplemental Fig. S6B). With the strong expression line pRPS5A>GR>EXPA1 (8-4), a statistically significant change in ν'Β was observed as soon as 3 h after EXPA1 induction (Fig. 4, D and E), similarly to long-term induction on media at both pH values (4 and 5.8). To exclude unspecific or side effects of gene overexpression, we determined the spatial map of cell stiffness using fluorescence emission–Brillouin imaging (Elsayad et al. 2016) in the Arabidopsis root over a larger area in a pEXPA15::EXPA15:mCherry line, revealing stronger fluorescent signal when compared with pEXPA1::EXPA1:mCherry. The regions of higher BFS associated well with the EXPA15 expression domain (Fig. 5), implying a possible role for EXPA15 in controlling CW stiffness. On the other hand, neither BLS nor AFM detected any change in CW stiffness in the root TZ of the expa1-2 knock-out line (Ramakrishna et al. 2019) when compared with WT (Supplemental Fig. S7).
Figure 4.
EXPA1 overexpression stiffens CWs measured via BFS. A) Representative images of 2D (xy) BFS maps in root cells of 7-d-old Arabidopsis seedlings of pRPS5A>GR>EXPA1 overexpressing EXPA1 (line 8-4) grown on MS media pH 5.8 +/− Dex. BFS expressed as Brillouin Elastic Contrast (ν'Β) was determined in roots of WT and the 8-4 line grown on solid MS media (B) +/− Dex pH 5.8, (C) +/− Dex pH 4, (D) grown on solid MS (without Dex) for 7 d and induced in Dex-supplemented liquid MS media pH 5.8 for 0.5 to 6 h, or (E) liquid MS media pH 4 for 0.5 to 6 h; DMSO was used in -Dex treatments. Medians shown are from at least 4 seedlings and 10 measurements in each category (n ≥ 10). All experiments were evaluated using simple ANOVA and post hoc Tukey test. Statistically significant differences at alpha 0.05 (*) and 0.001 (***) are shown. Scale bars in (A) correspond to 10 µm.
Figure 5.
EXPA15 localization overlaps with higher CW stiffness in the root tip as determined using fluorescence emission-Brillouin scattering. Fluorescence images (top raw, in red) and BFS (bottom raw, false color-coded) of 7-d-old Arabidopsis root from (A) WT and (B)pEXPA15::EXPA15:mCherry primary root and (C) emerging lateral root. The fluorescent signal corresponds to EXPA15 expression and overlaps with higher BFS. Scale bars correspond to 100 µm.
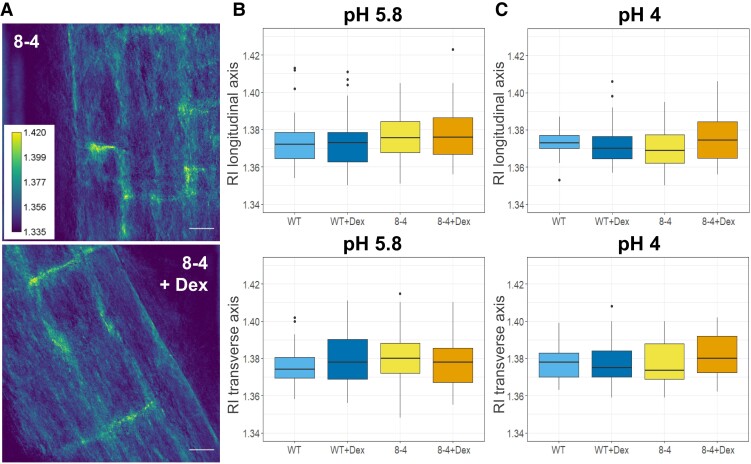
Since the refractive index (RI) in cells directly associates with the mass content, we applied quantitative cell tomography (using a holotomographic microscope) to measure the RI directly in the Arabidopsis roots in water. Representative maximum intensity projections of RI tomograms are shown in Fig. 6A. Quantitative data analysis confirmed that there are no statistically significant differences across all genotypes and treatment performed in both longitudinal (upper graphs) and transverse (lower graphs) CWs of the early elongating cells in Arabidopsis roots grown at both pH 4 and 5.8 (Fig. 6, B and C). Based on this finding, the effect of RI on the modulus calculation from the Brillouin shift can be expected to be minimal, supporting the use of ν'Β as a direct indicator of CW stiffness.
Figure 6.
RI measurements of Arabidopsis root CWs reveal comparable CW density in WT and EXPA1 overexpressing roots. A) RI tomograms (maximal projections) of root cells of 7-d-old Arabidopsis of seedlings pRPS5A>GR>EXPA1 overexpressing EXPA1 (line 8-4) grown on MS media pH 5.8 +/− Dex. Scale bars correspond to 20 μm. The graphs show RI measurements in water (RI 1.330) of roots of WT and the 8-4 line grown on MS media (B) +/− Dex pH 5.8 and (C) +/− Dex pH 4. Medians from a minimum of 6 seedlings and 30 measurements in each category (n ≥ 30) of longitudinal (upper) and transverse (lower) CW axes are shown. All experiments were evaluated using simple ANOVA and post hoc Tukey test; no statistically significant differences were observable (P > 0.1).
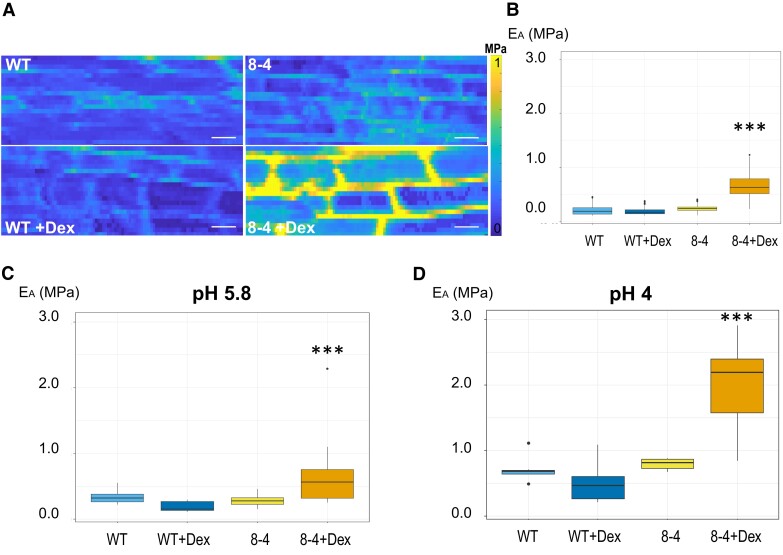
To directly measure the “stiffness” of root CWs, we used AFM. AFM-based microindentations apply precisely known forces on a cell through a cantilever and give a deformation value, from which we can extract Young's modulus of the cell (Peaucelle 2014, 2015). In complex plant tissues and given small deformations, the force to deform material is proportional to the area of indentation allowing the determination of a coefficient of proportionality that is named “apparent Young's modulus” (Peaucelle 2014). This coefficient depends on the speed of deformation and the mechanical characteristics of the sample. Representative heat maps of the apparent Young's modulus (EA) show clear differences when measured in the CW of root TZ/EZ boundary in 7-d-old Arabidopsis WT and EXPA1 overexpressing seedlings (Fig. 7A). The data confirm that the Dex-induced EXPA1 is associated with significantly stiffer root CW (P < 0.001) irrespective of whether stiffness is measured on anticlinal or periclinal CWs (Fig. 7, B and C, respectively) on growth media at either pH 5.8 or pH 4 (Fig. 7D). The stiffening effect of EXPA1 overexpression thus seems to be observable at indentation speed of the order of seconds (measured by AFM) and at GHz frequencies through the Brillouin technique.
Figure 7.
EXPA1 overexpression stiffens CWs when determined using AFM. A) Representative maps of the apparent Young's modulus (EA) of root cells of 7-d-old Arabidopsis WT and EXPA1 overexpressing seedlings (line 8-4) grown on MS media +/− Dex, showing differences in EA (representative of >50). The EA maps are presented as heat maps, with the respective scale, and show data from 2 successive maps of 60 × 80 and 60 × 80 force scans. Each pixel in the EA map represents the EA calculated from a single force-indentation curve, and each map consists of 4,800 data points. Images are 100 μm in length, scale bars correspond to 10 μm. B) Graphs present the EA of the anticlinal CW of roots as in (A) grown on MS media +/− Dex pH 5.8. Medians shown are from a minimum of 345 measurements in each category (n ≥ 345). C, D) Graphs present the EA of the periclinal CW of roots as in (A) grown on MS media +/− Dex at pH 5.8 (C) and pH 4 (D). The EA plotted on the graphs was determined by sampling data points within the area of interest. Medians shown are from a minimum of 6 measurements in each category (n ≥ 6). Statistically significant differences at alpha 0.001 (***) are shown. All experiments were evaluated using Mixed Model ANOVA and post hoc Tukey test. Statistically significant differences at alpha 0.001 (***) are shown.
To validate the ability of BLS to detect not only CW stiffening but also CW softening, we chose the well-described PECTIN METHYLESTERASE 5 overexpressor (PME5oe) lines that upon induction show substantially reduced stiffness of Arabidopsis epidermal hypocotyl cells due to pectin demethylesterification (Peaucelle et al. 2015). Both BLS and AFM revealed softer CW of PME5oe lines compared with the corresponding controls (Supplemental Fig. S8).
In sum, overexpression of EXPA1 results in CW stiffening measured at the TZ/EZ boundary using both BLS and AFM. Interestingly, even endogenous EXPA15 expression seems to be associated with cells displaying higher stiffness within the Arabidopsis root tip.
EXPA1 overexpression leads to CW remodeling
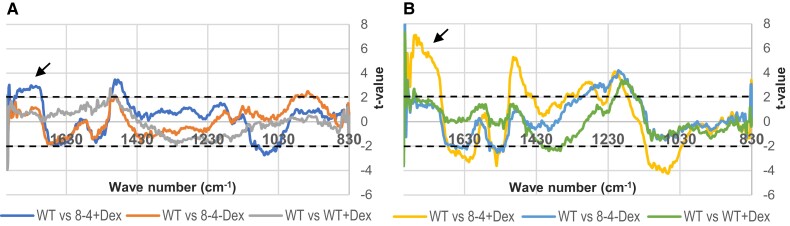
To get mechanistic insight into EXPA1 overexpression-induced CW stiffening, we chose to use a highly reproducible in situ Fourier-transform infrared (FT-IR) spectroscopy that provides a powerful and rapid assay for wall components and putative cross-links by identifying polymers and functional groups non-destructively (McCann et al. 1992; Mouille et al. 2003; Andres-Robin et al. 2018). FT-IR measurements (spectra 829 to 1,800 cm−1) were performed on the root TZ/early EZ of Arabidopsis WT and the EXPA1 overexpressing line. Dex induction was performed for a short 3 h period (Fig. 8A) or the seedlings were grown on Dex for a week (Fig. 8B). The most remarkable difference of absorbance observed at both time intervals (appearing after 3 h and being more pronounced after 7 d of EXPA1 upregulation; arrows in Figs. 8, A and B) could be assigned to ester linkages (around 1,730 cm−1) according to Mccann et al. (1992) and Mouille et al. (2003). This suggests a relatively higher esterification degree of pectin in the WT compared with the Dex-treated expansin overexpressor. Collectively, assaying CW composition via FT-IR shows that induction of EXPA1 expression leads to rapid (less than 3 h after induction) demethylesterification of pectin.
Figure 8.
EXPA1 overexpression changes physical–chemical properties of the CW as determined by FT-IR analysis of root CWs. FT-IR measurements of 7-d-old Arabidopsis WT and EXPA1 overexpressing seedlings (line 8-4) grown on MS media pH 5.8 without Dex (-Dex) and (A) induced with Dex (+Dex) for 3 h or (B) grown on Dex for 7 d. Root TZ/early EZ of 4 to 5 individual seedlings from at least 3 experiments (n > 12) were analyzed. Student's t-test of FT-IR spectra (829 to 1,800 cm−1) was performed to compare WT and the various treatments and genotypes. Graphs show the t-value (Y-axis) plotted against the wave numbers (x-axis). Horizontal dashed lines indicate the P = 0.95 significance threshold. The black arrows point towards 1730 cm−1.
To elucidate the mechanism underlying the observed EXPA1-induced CW remodeling, we assayed the genome-wide transcriptome changes both early (3 h) and later (7 d) after induction of EXPA1 overexpression. After 3 h of Dex induction, 336 differentially expressed genes (DEGs) were significantly upregulated and 287 downregulated when compared with non-induced controls (Dex-treated WT Col-0, Supplemental Data Set 1). Among the DEGs, significantly enriched gene ontology (GO) terms included plant-type CW loosening (GO0009828), modification (GO0009827), and organization (GO0009664), and apart from the intentionally overexpressed EXPA1 gene, pointed out a number of other expansin genes being misregulated (Supplemental Table S3). Of those, the most striking was a change in the transcriptional activity of EXPA2, reaching almost the fold change levels observed for Dex-induced EXPA1; however, the relatively smaller but still highly significant changes were observed also in the activity of several other expansin and expansin-like genes including EXPA4, EXPA8, EXPA15, EXPB3, and EXLB1 (Supplemental Table S4). GO term “CW organization or biogenesis” (GO0071554) comprising XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASEs (XTHs), particularly XTH5, XTH7, XTH11, and XTH32, was also found to be significantly enriched among the DEGs (Supplemental Table S3). After 7 d of Dex induction of the EXPA1 overexpressing line, only 127 genes were upregulated and 37 downregulated (compared with Dex-treated WT Col-0,Supplemental Data Set 1). Compared with the changes observed in the short-term response, apart from the significantly changed expression of EXPA1, EXPA2, and EXPA15, barely any changes in genes related to the CW processes were found (Supplemental Table S5). Dex caused no significant changes in CW modifications and only 15 and 20 genes were up- and downregulated, respectively.
To sum up, our data suggest that EXPA1 overexpression stimulates prompt changes in the expression of several CW modifying genes and associates with CW remodeling.
EXPA1 overexpression restrains root growth by reducing RAM size
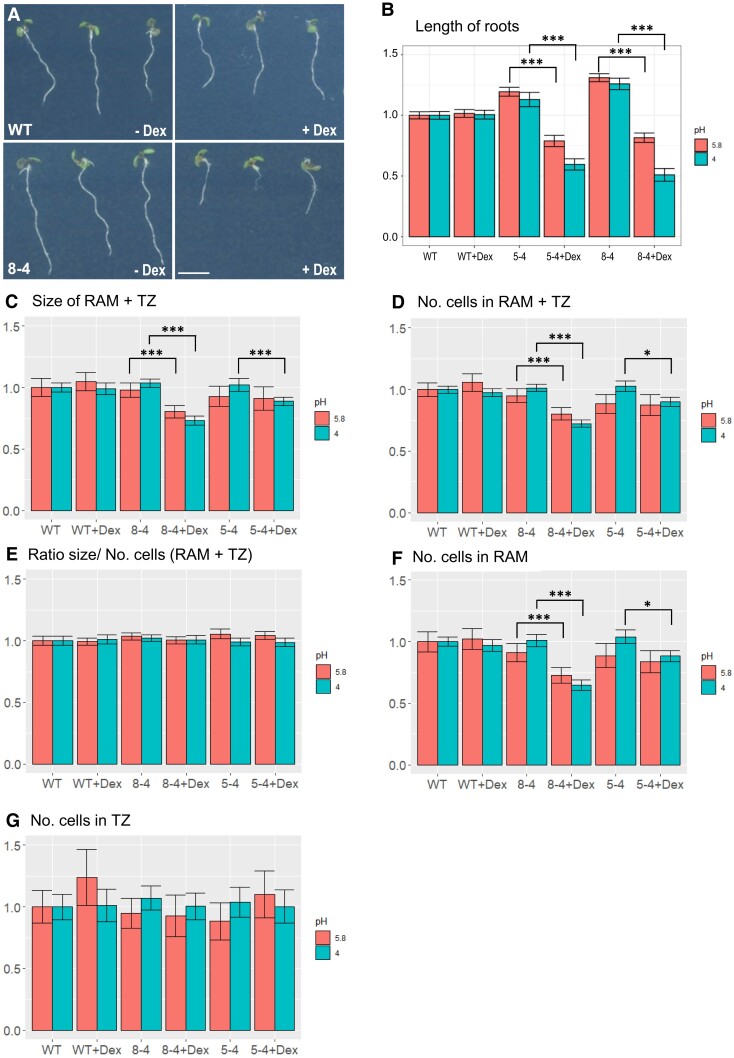
We examined the phenotype of WT and EXPA1 overexpressing seedlings (pRPS5A>GR>EXPA1 lines 5-4 and 8-4) grown on Dex continuously for a week. The Dex-induced plants had significantly reduced root length [by 25% to 30% (Figs. 9, A and B)]. The reduction was further enhanced to 40% to 73% when the pH of the growth media dropped from 5.8 to 4. To determine the possible mechanism of root shortening in more detail, we assayed the impact of EXPA1 overexpression on longitudinal root zonation as defined by Takatsuka et al. (2018). Our data show that RAM (meristematic zone/proximal meristem + TZ, Supplemental Fig. S9) size was significantly reduced by 18% and 29% for line 8-4 grown on media with normal (5.8) and acidic (4) pH, respectively (Fig. 9C). Similarly, cell number (counting in the cortex layer from the quiescent center to the first cell of the EZ) was significantly reduced (Fig. 9D). However, the RAM size/cell number ratio remained the same for each line with or without Dex induction (Fig. 9E), suggesting that the RAM cell number, but not cell length is reduced in the smaller roots. That was confirmed by a more detailed examination revealing that while the number of cells in the TZ remained similar to those of WT, the number of cells in the RAM proliferation zone (called the proximal meristem in Takatsuka et al. 2018) was significantly reduced upon EXPA1 overexpression (Fig. 9, F and G). Noteworthy, while the aforementioned effects on root length were significant in both lines (5-4 and 8-4) at both pH tested (5.8 and 4), the inhibitory effects on the RAM size in case of weaker overexpressor (line 5-4) were significant only at pH 4, suggesting acidic pH being beneficial for the EXPA1-mediated RAM shortening.
Figure 9.
EXPA1 overexpression reduces root growth by shortening the RAM. A) 7-d-old Arabidopsis seedlings of WT (top row) and pRPS5A>GR>EXPA1 line 8-4 (bottom row) grown on MS media pH4 supplemented either with DMSO (-Dex) or dexamethasone (+Dex). The scale bar is 5 mm. B) Length of roots, (C) the size of RAM + TZ, (D) the total number of cells (No. cells) in RAM + TZ, (E) the RAM size/cell number ratio, (F) No. of cells in RAM, and (G) No. of cells in TZ of 2 independent EXPA1 overexpressing lines (8-4 and 5-4) grown on MS media +/− Dex pH 5.8 and pH 4 relative to WT. Each experiment was repeated at least 3 times with a minimum of 10 seedlings in each category (n ≥ 30), and the error bars represent a 95% confidence interval. For all experiments, mixed model ANOVA using random effects for the different experiments was used with the Tukey test as a post hoc test. Statistically significant differences within genotypes and treatments at alpha 0.05 (*) and 0.001 (***) are shown.
In contrast to previous reports, in our hands, expa1-1 (Pacifici et al. 2018), expa1-2 (Ramakrishna et al. 2019), as well as our Dex-inducible amiRNA (amiEX1 lines, Supplemental Table S6), designed to downregulate EXPA1 and the closely related EXPA14 and EXPA15, did not display any significant phenotype in terms of root or RAM size (Supplemental Fig. S10). However, it should be noted here that following Dex induction, the amiEX1 lines only reduced EXPA1 expression by ∼50%, EXPA14 by 60%, and EXPA15 by 90% (RT-qPCR, Supplemental Fig. S11).
Taken together, while we do not see any effect of EXPA1 absence/downregulation on root growth and/or RAM size, the overexpression of EXPA1 results in fewer proliferating cells in the root meristem thus reduces Arabidopsis root growth.
Discussion
Is there a role for hormonal regulation of EXPA1 in root growth?
Recently, a role for Arabidopsis EXPA1 was proposed in the control of cell differentiation (expressed as a function of cell elongation) in cells leaving the meristematic zone of the RAM (Pacifici et al. 2018). Similar to a more recent study (Ramakrishna et al. 2019), our results do not show any statistically significant change in root length and/or RAM size in expa1-1, the CRISPR/Cas9 line expa1-2, or in our amiRNA lines. Nonetheless, it should be stressed here that exp1-2 is a hypomorphic allele (Ramakrishna et al. 2019) and our amiRNA lines are knock-down (not knock-out) lines. We did not detect EXPA1:mCherry outside the columella/LRC in the root tip and promoter activity (pEXPA1::nls:3xGFP) was only occasionally seen in the TZ/EZ boundary and in elongated cells proximal to that. In line with this, we did not detect any changes in CW stiffness in the root TZ of expa1-2 and no statistically significant upregulation of EXPA1 was detectable (using absolute fluorescence measurement) in the RAM using the pEXPA1::nls:3xGFP lines after 6 and 12 h treatment with both BAP and NAA. These findings suggest that the cytokinin-mediated transcriptional regulation of EXPA1 may take place in other parts of the root (e.g. cells surrounding LR primordia). Based on our data, we do not exclude a role for EXPA1 in controlling RAM size, but it probably does so in concert with other EXPAs (see below) and in the columella/LRC, where EXPA1 is active. The role of (cytokinin-regulated) auxin accumulation in the LRC in the control of RAM size has been proposed recently (Di Mambro et al. 2019). However, even with a more distinct transcriptional regulation of EXPA1 by NAA, we do not see any substantial and consistent EXPA1 upregulation in the LRC, both at the level of promoter activity and EXPA1 protein, thus leaving the functional importance of cytokinin- and auxin-mediated regulation over EXPA1 in the root tip rather unclear.
In planta expansin localization: specificity rather than redundancy
In previous studies, expansins were located in the CWs using immunolocalization techniques of fixed plant materials (Zhang and Hasenstein 2000; Cosgrove et al. 2002; Balestrini et al. 2005). Transgenic lines carrying EXPA genes in translational fusions with mCherry allowed observing the localization of EXPA proteins in living plants. Interestingly, our data suggest that the assayed EXPAs differ not only in the spatiotemporal specificity of expression, but their protein products also reveal a distinct localization pattern in specific domains of the root apoplast. Firstly, we see EXPA10 and EXPA14 localized predominantly in the longitudinal CWs of elongated root cells. This is similar to the situation observed in maize xylem, where the signal obtained after immunolocalization using anti-cucumber expansin antibody was homogenously distributed in isodiametric (non-elongated) xylem cells, while located mainly in the longitudinal CWs of elongated xylem (Zhang and Hasenstein 2000). Secondly, EXPA10, EXPA14, and EXPA15 (partially) showed a punctate pattern, spatially colocalizing with 3-cell boundaries, possibly in the surrounding intercellular space. These results imply the existence of a factor determining specific localization of individual EXPAs in different CW compartments. The homogenous distribution of EXPA1 throughout the CWs even outside its natural expression domain as seen in the pRPS5A>GR>EXPA1:mCherry line suggests that the specific localization pattern is not cell-type-specific, but rather encoded in the amino acid sequence of EXPA1. The existence and the molecular and/or biophysical nature of this factor responsible for homogenous expansin distribution in the CW and/or targeting a subset of EXPAs to specific apoplast domains remain elusive.
The Arabidopsis genome contains 26 genes for α-expansins (Li et al. 2002; Sampedro and Cosgrove 2005), suggesting functional diversification within the subfamily. Specific expression and localization of EXPA1, EXPA10, EXPA14, and EXPA15 together with differential hormonal sensitivity imply possible functional crosstalk among individual expansins. Overlap in their targeted action in the apoplastic continuum covering individual cells might result in the final vectorial change of CW expansion and the highly coordinated cellular behavior underlying root growth including its longitudinal zonation. A similar functional and spatiotemporal specificity including differential hormonal response and shoot cell growth-based zonation was described for LeEXP2, LeEXP9, and LeEXP18 in tomato (Caderas et al. 2000; Vogler et al. 2003). The functional importance and possible crosstalk and/or redundancy within the EXPA family in Arabidopsis, however, remains to be demonstrated.
Mechanism of EXPA-regulated CW biomechanics and root growth
Expansins were identified as factors facilitating pH-dependent CW loosening, allowing the cell to extend up to 100 times when compared with its length as a meristematic initial (see Cosgrove 2016 and references therein). To be able to do that, expansins are proposed to modify non-covalent bonds in the cellulose microfibril network, laterally interconnected via xyloglucans bound to the hydrophobic face of cellulose microfibrils (see Cosgrove 2018b and references therein). Consequently, fibril–fibril sliding seems to allow CW extension and in-plane stress release of the multi-lamellate CW structure (Zhang et al. 2019; Zhang et al. 2021). Importantly, expansin-controlled CW loosening allows in-plane stress relaxation and enlargement of CWs, but does not associate with changes in CW viscoelastic properties, as measured by tensile tests (Yuan et al. 2001; Cosgrove 2018a). Thus, the CW “stiffening” or “softening” (i.e. making the CW less or more deformable to out-of-plane mechanical force, respectively, measurable by, e.g. indentation techniques) is a mechanistically different process and should be distinguished from the EXPA-mediated CW loosening (Zhang et al. 2019; Samalova et al. 2022).
Using 2 independent approaches—non-invasive BLS imaging and AFM—we show here that overexpression of EXPA1, homogenously distributed throughout the CW, results in increased CW stiffness in root cells. Together with FT-IR analysis, these data suggest that besides its role in CW loosening, EXPA1 could be the modulator of CW viscoelastic properties by regulating CW remodeling. In general, EXPAs might modulate CW chemical composition by controlling the access of CW remodeling enzymes to their substrate either by relaxing CW structure (thus allowing different hydrolases to access their substrates) or by changing substrate availability due to EXPA binding to non-cellulose components of the CW matrix including hemicelluloses and pectin (McQueen-Mason and Cosgrove 1995; Georgelis et al. 2011; Wang et al. 2013). This was experimentally demonstrated for the putative carbohydrate-binding module (CBM) from strawberry expansin 2 (CBM-FaExp2). The binding of CBM-FaExp2 to CW carbohydrates was associated with a reduction in the activity of CW degrading enzymes including pectinase in the in vitro assays. Interestingly, CBM-FaExp2 shows a high level of similarity with AtEXPA1 and AtEXPA2 (Nardi et al. 2013). Thus, the high abundance of EXPA1 might interfere with the accessibility of pectin to pectin-modifying enzymes like methylesterases, pectin acetylesterases, polygalacturonases, or pectate-like lyases. This would potentially lead to pectin modifications and subsequent changes in CW rheological properties (Senechal et al. 2014). Our findings imply an interference between EXPA1 overexpression and methylesterification of pectin, previously demonstrated to be responsible for controlling CW viscoelasticity (reviewed in Levesque-Tremblay et al. 2015). Specifically, the FT-IR data suggest that EXPA1 is able to trigger rapid pectin demethylesterification that has been demonstrated to allow both CW stiffening and softening, possibly dependent on the demethylesterification pattern (Levesque-Tremblay et al. 2015). Potential functional artifacts due to high levels of EXPA1 overexpression used in our study cannot be completely excluded. Nonetheless, the ability of EXPA1 to induce CW stiffening as soon as 3 h after Dex addition (leading to an order of magnitude lower EXPA1 mRNA amount compared with the long-term induction) and an association between CW stiffness and EXPA15 expression (partially overlapping with EXPA1 expression pattern) in living Arabidopsis roots implicates expansins in the spatial-specific control of CW viscoelasticity, as proposed recently (Samalova et al. 2022). On one hand, it should be highlighted here that the co-localization of EXPA1 and EXPA15 with the CWs revealing higher relative stiffness in columella/LRC may be coincidental and does not provide evidence of any causal link between EXPA1/EXPA15 and CW stiffening. On the other hand, however, changes in the Raman spectra of pericycle CWs in expa1-1 provide strong evidence for the association of EXPA1 activity and CW composition in the EXPA1-dependent asymmetric pericycle (lateral root founder cell) expansion (Ramakrishna et al. 2019).
Besides the interference with pectin-modifying enzymes, another mechanism of EXPA1-induced CW remodeling could be EXPA1-mediated transcriptional regulation of CW modifying/loosening genes. By far the most upregulated one was EXPA2 (log2FC = 5.718). The gibberellic acid-inducible EXPA2 was shown to control endosperm expansion, being necessary for seed germination in Arabidopsis (Yan et al. 2014; Sanchez-Montesino et al. 2019). Interestingly, EXPA1 was found to be strongly downregulated in the mutant deficient in NAC25, the TF mediating the gibberellic acid-induced expression of EXPA2. Furthermore, the regulators mediating the GA-controlled endosperm expansion comprised also a number of genes we found to be misregulated by EXPA1 overexpression including several EXPAs (EXPA8, EXPA15) and XTHs (XTH5) (Sanchez-Montesino et al. 2019). This suggests that EXPA1 and EXPA2 together with several genes encoding for enzymes involved in the CW remodeling might be a part of the regulatory network involved in the (GA-regulated) cell elongation/expansion.
CW stiffness associated with upregulated EXPA1 seems to downregulate root growth by reducing RAM size. This suggests a mechanism connecting biomechanical CW properties with control over cell division in the RAM, as also implied by the previously demonstrated cell proliferation regulatory role for CW integrity (CWI) signaling (Gigli-Bisceglia et al. 2018). To maintain the functional CW, plants use specialized systems for monitoring and initiating compensatory responses often amplified through feedback processes. The sensors recognizing the CWI status and triggering the downstream transcriptional regulations are kinases from the Catharanthus roseus receptor-like kinase1-like (CrRLK1L) and Wall-associated kinase subfamilies. These kinases are able to interact with pectin, suggesting their possible role in sensing the disturbance in the CW matrix (see Baez et al. 2022 and references therein). CWI reacts to CW perturbation by changes in gene expression leading to CW deposition and remodeling as possible compensatory mechanisms (Voxeur and Hofte 2016; Baez et al. 2022). This implies time-dependent changes in the CWI-induced responses. In line with that, we see much stronger transcriptome changes early (3 h) after inducing EXPA1 OE (623 DEGs), while a much lower number of DEGs (164) was identifiable after the long-term (7 d) EXPA1 upregulation, thus hinting to the involvement of a mechanism mediating adaptation to EXPA1 overexpression. We may speculate on the involvement of CWI signaling-activated CW remodeling as a possible compensatory mechanism responding to CW disturbance and allowing to reach the new equilibrium (see Vaahtera et al. 2019; Samalova et al. 2022 and references therein).
BLS vs. AFM
In terms of the methodological approach used, there are important differences between the longitudinal elastic modulus (M) measured by BLS and Young's modulus (E) measured via AFM (Prevedel et al. 2019). The BLS measured M is well known to be very sensitive to the level of hydration (Palombo et al. 2014; Wu et al. 2018; Andriotis et al. 2019) and temperature (Berne and Pecora 2000), and any comparisons thus need to be made under the same thermodynamic conditions and hydration levels. However, as these can be assumed to be similar between the different samples measured, variations between samples can be interpreted as being due to changes in the mechanical properties in the probed regime. The comparable trend of the quasi-static Young modulus (measured by AFM) and BLS measured ν'Β observed here is consistent with observations in a diverse set of biological samples (e.g. Scarcelli et al. 2015; Andriotis et al. 2019; Gouveia et al. 2019) suggesting that here too the latter may serve as a proxy for stiffness. We note, however, that at very high hydration levels (much higher than in the system studied here), the relation between the 2 can be expected to break down (Wu et al. 2018).
Conclusions
Based on our and others’ results, we propose an important regulatory role in controlling root growth and development in Arabidopsis for the tightly controlled spatiotemporal specificity of expansin expression and the localization of their protein products into distinct domains of the extracellular matrix, possibly in conjunction with hormone-regulated pH distribution in the root apoplast (Barbez et al. 2017; Pacifici et al. 2018; Großeholz et al. 2021; Li et al. 2021; Serre et al. 2022). We also propose that apart from mediating CW loosening, expansins may control viscoelastic properties of CWs by regulating CW remodeling. That implicates expansins as important modulators of plant development controlling CW biomechanical properties through a complex regulatory mechanism.
Materials and methods
Plant cultivation conditions and dexamethasone (Dex) induction
Standard MS medium supplemented with 1.5% (w/v) sucrose, 0.8% (w/v) plant agar (Duchefa), and pH 5.8 (adjusted with KOH) or pH 4 (adjusted with H2SO4) was used. Arabidopsis (A. thaliana) plants were cultivated in growth chambers under long-day conditions (16 h light/8 h dark) at 21 °C in Petri dishes or in soil, with a light intensity of 150 μM m−2 s−1 and 40% relative humidity. Dex induction was performed by adding 20 μM Dex into MS media on plates; for short time exposure (1 to 6 h or 24 h), seedlings were placed into liquid MS media containing 20 μM Dex (as described in Samalova et al. 2019). DMSO at the same concentration was used instead of Dex as a control.
Confocal laser scanning microscopy and image analysis
We used a Zeiss LSM 880 laser scanning microscope to localize EXPA:mCherry fusions. mCherry fluorescence was detected at 580 to 650 nm with 561-nm HeNe laser excitation and eGFP at 490 to 550 nm with a 488-nm Argon laser line with a detector gain set to 800. Z-stack projections are shown at maximum intensity. Fluorescence quantification was done using CellProfiler (mCherry) and Imaris (nls:3xGFP) software. To measure RAM size, 7-d-old Arabidopsis seedlings were stained with 30 μg ml−1 propidium iodide for 5 min, scanned at 590 to 650 nm with 488-nm excitation, and measured using ZEN 3.0 software. The roots were imaged using the 40×/1.2 water-corrected C-Apochromat or the 25×/0.8 immersion-corrected Plan-Apochromat objectives.
BLS microscopy
BLS is the inelastic scattering of light from inherent or stimulated high-frequency acoustic vibrations in a sample, the speed of which is directly related to the elastic modulus of the material (Berne and Pecora 2000). The BFS (νΒ) is proportional to the acoustic phonon velocity, which is in turn proportional to the square root of the high-frequency longitudinal elastic modulus (M). As such, the Brillouin frequency can serve as a proxy for the mechanical properties of the sample. In particular, M is closely related to the compressibility of the sample and has empirically been observed to scale semi-logarithmically with Young's modulus (E) as measured by AFM in diverse samples including live cells (Scarcelli et al. 2015). An accurate calculation of M requires knowledge of the ratio n2/ρ , where n and ρ are the RI and the mass density, respectively, in the probed region of the sample. While it can often be assumed by virtue of the Lorentz–Lorenz (LL) relation that n2 will scale with ρ (Zhao et al. 2011), such that explicit knowledge of the ratio n2/ρ at each probed region is not required, the validity of the LL-relation in complex multicomponent structures including the CW cannot be rigorously justified. As such, we present results in terms of a dimensionless frequency shift previously introduced as the Brillouin Elastic Contrast, ν'Β = νΒ /νΒ(w)−1, where νΒ(w) is the measured BLS frequency of distilled water (Antonacci et al. 2020). As is the case for the BFS, ν'Β scales with the square root of M. Moreover, it is independent of the probing wavelength; it can correct for slight variations in temperature between measurements and allows for more direct comparisons of measurements between instruments employing different probing wavelengths.
Brillouin microscopy was performed using the in-house Brillouin confocal microscope described in Elsayad et al. (2016). Excitation was via a single-mode 532 nm laser (Torus, Laser Quantum, DE). A dual cross-dispersion Virtual Imaged Phase Array (VIPA) spectrometer (Scarcelli et al. 2015) with a Lyott Stop (Edrei et al. 2017) was used to measure BLS spectra. The spectral projection was measured on a cooled EM CCD camera (ImageEMX II, Hamamatsu, JP). The spectrometer was coupled to an inverted microscope frame (IX73, Olympus, JP) via a physical pinhole with an effective size of 1 Airy Unit to ensure optimum confocal detection. After the pinhole, a dichroic mirror was used to outcouple light with wavelengths longer than 536 nm to a fluorescence spectrometer (Ocean Optics QE Pro, USA) to detect the fluorescence signal assuring pixel-to-pixel association with the measured Brillouin spectra. To acquire Brillouin maps, samples were scanned in x, y, and/or z using either a 3-axis long-range Piezo-stage (Physik Instrumente, DE) or a motor stage (ASI, USA), both mounted on top of the inverted microscope frame. Light could also be coupled out through a second port on the microscope frame using a long-pass filter (AHF, DE) and a tube lens to a compact sCMOS camera (Thorlabs, DE) allowing us to locate samples and regions of interest (in wide-field transmitted-light conditions illuminating the sample from the top with a halogen lamp) as well as to monitor the position being probed during scanning.
All hardware was controlled using LabView (National Instruments, USA)-based software developed by THATec (DE), especially for our microscope. The 16-bit depth spectral projection image for each position in the spatial scan was exported from the native THATec format into MATLAB (Mathworks, DE), where a custom-written code was used for analysis. This code (see also Elsayad et al. 2016) used 2 calibration spectra (of triple distilled water and spectroscopic grade ethyl alcohol) measured before and after each set of scans. These were used for registration of the spectral projection onto a frequency scale, based on the calculated dispersion for a dual-VIPA setup in the paraxial approximation regime. The alignment of the spectrometer was such that maximal energy was transferred into a single diffraction order. Due to the spatial masking of the elastic scattering peaks at the 2 intermediate imaging planes in the spectrometer, the spectral projection consisted of only 2 inelastic scattering peaks corresponding to the so-called Brillouin Stokes and anti-Stokes scattering peaks.
All data analysis was performed in MATLAB (Mathworks, DE) using custom-written scripts (Elsayad et al. 2016). Spectral phasor analysis (Elsayad 2019) was used to obtain initial parameter estimates for peak positions and widths which were subsequently inserted into a non-linear least squares fitting algorithm that fitted 2 broadened Lorentzian functions (Voigt functions) to obtain the 2 peak positions, from which the BFS could be obtained. The BLS spectra were also deconvolved in phasor space using a response function obtained from measuring the attenuated Rayleigh scattering inside the respective samples (by opening the spatial masks).
For all scans, the laser power at the sample was between 1 and 5 mW, and the dwell time per point, which was also the acquisition time of each spectrum, was 100 ms. Cells were inspected (by transmitted-light wide-field illumination) and appeared healthy and unperturbed after the experiments, suggesting that BLS measurements had no negative or phototoxic effects. A 1.4 NA objective was used for excitation and detection (back-scattering geometry). As such a broad range of scattering wavevectors is probed, and one effectively probes directionally averaged elastic moduli. As a direct consequence of probing a broad spectrum of wavevectors, the Brillouin spectra are broadened as predicted from the momentum-energy conservation equations describing the scattering processes. The so-called Brillouin scattering peak position is, however, not noticeably modified to within experimental uncertainties, as was verified by reducing the numerical aperture of excitation and detection on the studied samples using an iris in the beam path.
Roots of 7-d-old Arabidopsis seedlings were scanned at the early EZ, the size of the scan was 25 µm × 25 µm (50 × 50 pixels), with step size typically 500 nm (or 250 nm for larger scans) using the piezo-stage.
RI tomography
RI tomograms were acquired on a holotomographic microscope with rotational scanning 3D Cell Explorer (Nanolive SA, Lausanne, Switzerland) with Nikon BE Plan 60× NA 0.8. The size of the acquired tomogram was 93.1 × 93.1 × 35.7 μm (xyz). Samples were measured in water (reference RI 1.330). Steve 1.6.3496 software (Nanolive SA) was used for image acquisition. Subsequent image analysis was performed in ImageJ 1.52q (NIH, USA) on a max projection of tomography data. The median RI at the CW of the longitudinal and transverse axes of the cell was then extracted.
Atomic force microscopy
Roots of 7-d-old Arabidopsis seedlings were immobilized on glass slides and surrounded by stiff agarose. Approximate TZ/early EZ was defined based on the visual landmark observed through a bright field microscope. In order to extract the mechanical properties of only the outer CW, the maximum indentation force was set to 60 nN to archive a maximum indentation of no more than 80 nm. The cantilever used was “Nano World” (Nanosensors Headquarters, Neuchâtel, Switzerland) SD-R150-T3L450B tips with a spring constant of 0.15 to 1.83 N/m (the one used was estimated to be 0.781 N/m) with silicon point probe tips with a 150-nm radius.
In order to extract the mechanical properties of the periclinal CW, the maximum indentation force was set to 70 nN to achieve a maximum indentation of no more than 80 nm. The cantilever used was “Nano World” (Nanosensors Headquarters, Neuchâtel, Switzerland) SD-Sphere-FM-S-10 tips with a spring constant of 0.15 to 1.83 N/m (the one used was estimated to be 1.1 N/m) with silicon point probe tips with an 800-nm radius.
All force spectroscopy experiments were performed as previously described (Peaucelle 2014; Peaucelle et al. 2015; Feng et al. 2018). Briefly, the stiffness of samples was determined as follows: an AFM cantilever loaded with a spherical tip was used to indent the sample over a 60 × 100 μm square area, within the area 60 × 100 measurements were made resulting in 6000 force-indentation experiments; each force-indentation experiment was treated with a Hertzian indentation model to extract the apparent Young modulus (EA); each pixel in the stiffness map represents the apparent Young modulus from 1 force-indentation point. The EA was calculated using JPK Data Processing software (ver. Spm-4.0.23, JPK Instruments AG, Germany), which allows for a more standardized analysis than the estimation of the EA using a standard Hertzian contact model (Peaucelle 2014, 2015). As is typically the case in nano-indentation experiments, only the retraction curve was used in our analyses. A Poisson ratio of 0.5 was assumed for the material. Range distribution of EA from 0.2 to 3 MPa in 1-MPa binned groups was calculated using MATLAB. The topography map generated from the contact point calculation permitted the selection of the periclinal or anticlinal CW perpendicular to the indentation for the analysis.
FT-IR analysis
Seven-day-old Arabidopsis seedlings of WT and EXPA1 overexpressing line (8-4) grown on MS media pH 5.8 +/− Dex were collected and stored in ethanol. They were then rehydrated in water overnight prior to being laid out on gold-coated slides and left to dry for a few hours at room temperature. Spectra were collected using a Nicolet iN10 Infrared Imaging Microscope (Thermofisher) in a 30 × 30 µm window. Root TZ of 4 to 5 individual seedlings from at least 3 experiments (n > 12) were analyzed for each treatment/genotype. Eight interferograms were collected in transmission mode with 8 cm−1 resolution and coadded to improve signal-to-noise ratios. The collected spectra were baseline corrected and normalized using previously described procedures (Mouille et al. 2003).
Statistical analysis
For statistical analyses, simple ANOVA and post hoc Tukey test were used. For pairwise comparisons in repeated experiments, mixed model ANOVA using random effects for the different experiments was used with the Tukey test as a post hoc test. In the case of non-normal count data (e.g. No. of cells), a Poisson mixed model was used to identify differences between genotypes. The lme4 package in R was used for the implementation of the mixed models (Bates et al. 2015).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: EXPA1 (At1g69530), EXPA10 (At1g26770), EXPA14 (At5g56320), and EXPA15 (At2g03090).
Supplementary Material
Acknowledgments
We thank Ive De Smet for his kind donation of the pEXP1::nls:3xGFP seeds, Alexis Maizel for CRISPR/Cas9 exp1-2 Arabidopsis lines, and Victoria Mironova for critical reading of the manuscript.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Marketa Samalova, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic; Department of Experimental Biology, Faculty of Science, Masaryk University, Brno 625 00, Czech Republic.
Alesia Melnikava, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic; National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Brno 625 00, Czech Republic.
Kareem Elsayad, Division of Anatomy, Centre for Anatomy & Cell Biology, Medical University of Vienna, Vienna 1090, Austria.
Alexis Peaucelle, INRAE, Versailles-Grignon 78026, France.
Evelina Gahurova, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic; National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Brno 625 00, Czech Republic.
Jaromir Gumulec, Department of Pathological Physiology, Faculty of Medicine, Masaryk University, Brno 625 00, Czech Republic.
Ioannis Spyroglou, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic.
Elena V Zemlyanskaya, Department of Natural Sciences, Novosibirsk State University, Novosibirsk 630073, Russia; Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk 630090, Russia.
Elena V Ubogoeva, Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk 630090, Russia.
Darina Balkova, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic; Department of Experimental Biology, Faculty of Science, Masaryk University, Brno 625 00, Czech Republic.
Martin Demko, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic.
Nicolas Blavet, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic.
Panagiotis Alexiou, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic.
Vladimir Benes, Genomics Core Facility, European Molecular Biology Laboratory, Heidelberg 69117, Germany.
Gregory Mouille, INRAE, Versailles-Grignon 78026, France.
Jan Hejatko, CEITEC – Central European Institute of Technology, Masaryk University, Brno 625 00, Czech Republic; National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Brno 625 00, Czech Republic.
Author contributions
M.S. and J.H. designed the research, M.S., A.M., K.E., A.P., E.G., J.G., E.V.Z., E.V.U., D.B., V.B., and G.M. performed the research, M.S., K.E., A.P., J.G., I.S., E.V.Z., M.S., N.B., P.A., V.B., G.M., and J.H. analyzed the data, and M.S., K.E., A.P., J.G., G.M., and J.H. wrote the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. EXPA1 is not inducible by cytokinins in the root tip, and cytokinin-responsive transcription factors bind to EXPA promoters.
Supplemental Figure S2. EXPA1 activity is only occasionally detectable in the root TZ, while EXPA1 signal is delimited to the columella/lateral root cap.
Supplemental Figure S3. Cell-type-specific localization of EXPA14, EXPA15, and EXPA10 that localizes predominantly to the longitudinal cell walls.
Supplemental Figure S4. Confocal imaging of a plasma membrane marker line UBQ10::YFP-PIP1;4 before and after plasmolysis.
Supplemental Figure S5. Low-level EXP1A:mCherry overexpression does not reveal cell wall stiffening, as determined using Brillouin light scattering microscopy and RT-qPCR of Dex-induced levels of expression of EXP1A:mCherry and EXPA1 in selected lines.
Supplemental Figure S6. Short-time Dex induction is sufficient to increase EXPA1 expression, as detected by RT-qPCR and EXPA1:mCherry fluorescence.
Supplemental Figure S7. expa1-2 knock-out does not show changes in cell wall stiffening in the TZ, as determined using Brillouin light scattering microscopy and atomic force microscopy.
Supplemental Figure S8. Comparison of BLS and AFM measurements on PME5oe overexpressing seedlings.
Supplemental Figure S9. An example of CLSM images used to determine the size of RAM.
Supplemental Figure S10. Root growth is not affected in expa1 knock-out and EXPA knock-down lines.
Supplemental Figure S11. RT-qPCR of Dex-induced reduction of expansin expression in amiEX1lines.
Supplemental Materials and Methods .
Supplemental Table S1. List of primers used for RT-qPCR.
Supplemental Table S2. List of primers used for expansin cloning.
Supplemental Table S3. GO terms significantly enriched among DEGs identified 3 h after Dex-induced EXPA1 overexpression.
Supplemental Table S4. CW-related DEGs identified 3 h after Dex-induced EXPA1 overexpression.
Supplemental Table S5. CW-related DEGs identified 7 d after Dex-induced EXPA1 overexpression.
Supplemental Table S6. List of primers used for cloning of amiEX1.
Supplemental References.
Supplemental Data Set 1. DESeq2 data analysis.
Funding
M.S. has received funding from the Czech Science Foundation, project No. 22-17501S. The work was further supported by the Ministry of Education, Youth and Sports of CR from the European Regional Development Fund-Project “Centre for Experimental Plant Biology”: No. CZ.02.1.01/0.0/0.0/16_019/0000738, LTAUSA18161 and the Czech Science Foundation (19-24753S). K.E. acknowledges support from the City of Vienna and the Austrian Ministry of Science (Vision 2020). The work of E.V.Z. and E.V.U. was supported by the Russian Science Foundation (20-14-00140) and the Russian State Budgetary Project (FWNR-2022-0006). We acknowledge Plant Sciences, Bioinformatics, and Cellular Imaging CFs at CEITEC MU supported by MEYS CR (LM2018129).
REFERENCES
- Andres-Robin A, Reymond MC, Dupire A, Battu V, Dubrulle N, Mouille G, Lefebvre V, Pelloux J, Boudaoud A, Traas J, et al. . Evidence for the regulation of gynoecium morphogenesis by ETTIN via cell wall dynamics. Plant Physiol. 2018:178(3):1222–1232. 10.1104/pp.18.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriotis OG, Elsayad K, Smart DE, Nalbach M, Davies DE, Thurner PJ. Hydration and nanomechanical changes in collagen fibrils bearing advanced glycation end-product. Biomed Opt Express. 2019:10(4):1841–1855. 10.1364/BOE.10.001841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci G, Beck T, Bilenca A, Czarske J, Elsayad K, Guck J, Kim K, Krug B, Palombo F, Prevedel R, et al. . Recent progress and current opinions in Brillouin microscopy for life science applications. Biophys Rev. 2020:12(3):615–624. 10.1007/s12551-020-00701-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacete L, Schulz J, Engelsdort T, Bartosova Z, Vaahtera L, Zan G, Gerhold JM, Ticha T, Ovstebo C, Gigli-Bisceglia N, et al. . THESEUS1 Modulates cell wall stiffness and abscisic acid production in Arabidopsis thaliana. Proc Natl Acad Sci. 2022:119(1):e2119258119. 10.1073/pnas.2119258119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez LA, Ticha T, Hamann T. Cell wall integrity regulation across plant species. Plant Mol Biol. 2022:109(4–5):1–22. 10.1007/s11103-022-01284-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R, Cosgrove DJ, Bonfante P. Differential location of alpha-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta. 2005:220(6):889–899. 10.1007/s00425-004-1431-2 [DOI] [PubMed] [Google Scholar]
- Barbez E, Dunser K, Gaidora A, Lendl T, Busch W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc Natl Acad Sci. 2017:114(24):E4884–E4893. 10.1073/pnas.1613499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015:67(1):1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Berne BJ, Pecora R. Dynamic light scattering, with applications to chemistry, biology, and physics. New York: (NY: ): Dover Publications; 2000. [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang Y-H, Schaller GE, Loraine A, Kieber JJ. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013:162(1):272–229. 10.1104/pp.113.217026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook S, Jönsson H. Shifting foundations: the mechanical cell wall and development. Curr Opin Plant Biol. 2016:29:115–120. 10.1016/j.pbi.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JKC, McQueen-Mason S, Kuhlemeier C. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol. 2000:123(4):1399–1413. 10.1104/pp.123.4.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002:14(12):3237–3253. 10.1105/tpc.006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T, Kende H. Tissue localization of expansins in deepwater rice. Plant J. 1998:15(6):805–812. 10.1046/j.1365-313X.1998.00258.x [DOI] [PubMed] [Google Scholar]
- Cleland R. Cell wall extension. Ann Rev Plant Physiol. 1971:22(1):197–222. 10.1146/annurev.pp.22.060171.001213 [DOI] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005:6(11):850–861. 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol. 2014:22:122–131. 10.1016/j.pbi.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Catalysts of plant cell wall loosening. F1000Research. 2016:5:119. 10.12688/f1000research.7180.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Nanoscale structure, mechanics and growth of epidermal cell walls. Curr Opin Plant Biol. 2018a:46:77–86. 10.1016/j.pbi.2018.07.016 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Diffuse growth of plant cell walls. Plant Physiol. 2018b:176(1):16–27. 10.1104/pp.17.01541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho H-T, Hoffmann-Benning S, Moore RC, Blecker D. The growing world of expansins. Plant Cell Physiol. 2002:43(12):1436–1444. 10.1093/pcp/pcf180 [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 2005:41(6):899–918. 10.1111/j.1365-313X.2005.02342.x [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Sabatini S. Emerging role of cytokinin as a regulator of cellular differentiation. Curr Opin Plant Biol. 2008:11(1):23–27. 10.1016/j.pbi.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007:17(8):678–682. 10.1016/j.cub.2007.02.047 [DOI] [PubMed] [Google Scholar]
- Didi V, Jackson P, Hejatko J. Hormonal regulation of secondary cell wall formation. J Exp Bot. 2015:66(16):5015–5027. 10.1093/jxb/erv222 [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, et al. . Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci. 2017:114(36):E7641–E7649. 10.1073/pnas.1705833114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, Svolacchia N, Dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, et al. . The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol. 2019:29(7):1199–1205. 10.1016/j.cub.2019.02.022 [DOI] [PubMed] [Google Scholar]
- Edrei E, Gather MC, Scarcelli G. Adaptive optics in spectroscopy and densely labeled-fluorescence applications. Opt Express. 2017:26(26):33865–33877. 10.1364/OE.26.033865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayad K. Spectral phasor analysis for Brillouin microspectroscopy. Front Phys. 2019:7:62. 10.3389/fphy.2019.00062 [DOI] [Google Scholar]
- Elsayad K, Polakova S, Gregan J. Probing mechanical properties in biology using Brillouin microscopy. Trends Cell Biol. 2019:8(8):608–611. 10.1016/j.tcb.2019.04.002 [DOI] [PubMed] [Google Scholar]
- Elsayad K, Werner S, Gallemí M, Kong J, Sánchez Guajardo ER, Zhang L, Jaillais Y, Greb T, Belkhadir Y. Mapping the subcellular mechanical properties of live cells in tissues with fluorescence emission–Brillouin imaging. Sci Signal. 2016:435:rs5. doi: 10.1126/scisignal.aaf6326 [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006:126(4):677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. . The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol. 2018:28(5):666–675. 10.1016/j.cub.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997:276(5317):1415–1418. 10.1126/science.276.5317.1415 [DOI] [Google Scholar]
- Geitmann A, Ortega JKE. Mechanics and modeling of plant cell growth. Trends Plant Sci. 2009:14(9):467–478. 10.1016/j.tplants.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Georgelis N, Tabuchi A, Nikolaidis N, Cosgrove DJ. Structure-function analysis of the bacterial expansin EXLX1. J Biol Chem. 2011:286(19):16814–16823. 10.1074/jbc.M111.225037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, Engelsdorf T, Strnad M, Vaahtera L, Khan GA, Yamoune A, Alipanah L, Novak O, Persson S, Hejatko J, et al. . Cell wall integrity modulates Arabidopsis thaliana cell cycle gene expression in a cytokinin- and nitrate reductase-dependent manner. Development. 2018:145(19):dev166678. 10.1242/dev.166678 [DOI] [PubMed] [Google Scholar]
- Gouveia RM, Lepert G, Gupta S, Mohan RR, Paterson C, Connon CJ. Assessment of corneal substrate biomechanics and its effect on epithelial stem cell maintenance and differentiation. Nat Commun. 2019:10(1):1496. 10.1038/s41467-019-09331-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großeholz R, Wanke F, Glöckner N, Rausch L, Rohr L, Scholl S, Scacch E, Spaziere A-J, Shabala L, Shabala S, et al. . Computational modeling and quantitative cell physiology reveal central parameters for the brassinosteroid-regulated cell growth of the Arabidopsis root. Elife. 2022:11. 10.7554/eLife.73031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruel J, Landrein B, Tarr P, Schuster C, Refahi Y, Sampathkumar A, Hamant O, Meyerowitz EM, Jonsson H. An epidermis-driven mechanism positions and scales stem cell niches in plants. Sci Adv. 2016:2(1):e1500989. 10.1126/sciadv.1500989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Meyerowitz EM, Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science. 2020:367(6481):1003–1007. 10.1126/science.aaz5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A, Menzel H, Krauss A. Versuche und Hypothese zur Primarwirkung des Auxin beim Streckungswachtum. Planta. 1971:100(1):47–75. 10.1007/BF00386886 [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. . Developmental patterning by mechanical signals in Arabidopsis. Science. 2008:322(5908):1650–1655. 10.1126/science.1165594 [DOI] [PubMed] [Google Scholar]
- Hamant O, Traas J. The mechanics behind plant development. New Phytol. 2010:185(2):369–385. 10.1111/j.1469-8137.2009.03100.x [DOI] [PubMed] [Google Scholar]
- Hervieux N, Tsugawa S, Fruleux A, Dumond M, Routier-Kierzkowska AL, Komatsuzaki T, Boudaoud A, Larkin JC, Smith RS, Li CB, et al. . Mechanical shielding of rapidly growing cells buffers growth heterogeneity and contributes to organ shape reproducibility. Curr Biol. 2017:27(22):3468–3479. 10.1016/j.cub.2017.10.033 [DOI] [PubMed] [Google Scholar]
- Hurny A, Cuesta C, Cavallari N, Otvos K, Duclercq J, Dokladal L, Montesinos JC, Gallemi M, Semeradova H, Rauter T, et al. . SYNERGISTIC ON AUXIN AND CYTOKININ 1 positively regulates growth and attenuates soil pathogen resistance. Nat Commun. 2020:11(1):2170. 10.1038/s41467-020-15895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al. . Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. eLife. 2015:4: e07811. 10.7554/eLife.07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013:54(10):1600–1611. 10.1093/pcp/pct105 [DOI] [PubMed] [Google Scholar]
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J. (2007) Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol Genet Genomics 277:115–137. [DOI] [PubMed] [Google Scholar]
- Levesque-Tremblay G, Pelloux J, Braybrook SA, Muller K. Tuning of pectin methylesterification: consequences for cell wall biomechanics and development. Planta. 2015:242(4):791–781. 10.1007/s00425-015-2358-5 [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 2002:128(3):854–864. 10.1104/pp.010658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Verstraeten I, Roosjen M, Takahashi K, Rodriguez L, Merrin J, Chen J, Shabala L, Smet W, Ren H, et al. . Cell surface and intracellular auxin signalling for H+ fluxes in root growth. Nature. 2021:599(7884):273–277. 10.1038/s41586-021-04037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Strauss S, Adibi M, Mosca G, Yoshida S, Dello Ioio R, Runions A, Andersen TG, Grossmann G, Huijser P, et al. . Cytokinin promotes growth cessation in the Arabidopsis root. Curr Biol. 2022:32(9):1–12. 10.1016/j.cub.2022.03.019 [DOI] [PubMed] [Google Scholar]
- Majda M, Grones P, Sintorn IM, Vain T, Milani P, Krupinski P, Zagorska-Marek B, Viotti C, Jonsson H, Mellerowicz EJ, et al. . Mechanochemical polarization of contiguous cell walls shapes plant pavement cells. Dev Cell. 2017:43(3):290–300. 10.1016/j.devcel.2017.10.017 [DOI] [PubMed] [Google Scholar]
- McCann MC, Hammouri M, Wilson R, Belton P, Roberts K. Fourier Transform infrared microspectroscopy is a new way to look at plant cell walls. Plant Physiol. 1992:100(4):1940–1947. 10.1104/pp.100.4.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Expansin mode of action on cell walls: analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995:107(1):87–100. 10.1104/pp.107.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992:4:1425–1433. doi: 10.1105/tpc.4.11.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform Infrared (FT-IR) microspectroscopy. Plant J. 2003:35(3):393–404. 10.1046/j.1365-313X.2003.01807.x [DOI] [PubMed] [Google Scholar]
- Nardi C, Escudero C, Villarreal N, Martinez G, Civello PM. The carbohydrate-binding module of Fragaria x ananassa expansin 2 (CBM-FaExp2) binds to cell wall polysaccharides and decreases cell wall enzyme activities “in vitro”. J Plant Res. 2013:126(1):151–159. 10.1007/s10265-012-0504-8 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016:165(5):1280–1292. 10.1016/j.cell.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E, Di Mambro R, Dello Ioio R, Costantino P, Sabatini S. Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J. 2018:37(16):e99134. 10.15252/embj.201899134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo F, Winlove CP, Edginton RS, Green E, Stone N, Caponi S, Madami M, Fioretto D. Biomechanics of fibrous proteins of the extracellular matrix studied by Brillouin scattering. J R Soc Interface. 2014:11(101):20140739. 10.1098/rsif.2014.0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 2012:158(1):465–475. 10.1104/pp.111.189779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Li F, Renaud J, Shen W, Li Y, Guo L, Cui H, Sumarah M, Wang A. NbEXPA1, an (-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 2017:92(5):846–886. 10.1111/tpj.13723 [DOI] [PubMed] [Google Scholar]
- Peaucelle A. AFM-based mapping of the elastic properties of cell walls: at tissue, cellular, and subcellular resolutions. J Vis Exp. 2014:89:51317. doi: 10.3791/51317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011:21(20):1720–1726. 10.1016/j.cub.2011.08.057 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol. 2008:18(24):1943–1948. 10.1016/j.cub.2008.10.065 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Höfte H. The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol. 2015:25(13):1746–1752. 10.1016/j.cub.2015.05.022 [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci. 2001:98(20):11812–11817. 10.1073/pnas.191380498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedel R, Diz-Munoz A, Ruocco G, Antonacci G. Brillouin microscopy: an emerging tool for mechanobiology. Nat Methods. 2019:16(10):969–977. 10.1038/s41592-019-0543-3 [DOI] [PubMed] [Google Scholar]
- Ramakrishna P, Ruiz Duarte P, Rance GA, Schubert M, Vordermaier V, Dai Vue L, Murphy E, Vilches Barro A, Swarup K, Moirangthema K, et al. . EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc Natl Acad Sci. 2019:116(17):8597–8602. 10.1073/pnas.1820882116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998:10(9):1427–1437. 10.1105/tpc.10.9.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, de Silva NV, Santos T, Abrantes F, Custodio C, Machado Neto N, Vieira L. Regulation of α-expansins genes in Arabidopsis thaliana seeds during post-osmopriming germination. Physiol Mol Biol Plants. 2019:25(2):511–522. 10.1007/s12298-018-0620-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M, Brzobohaty B, Moore I. Pop6/LhGR: a stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J. 2005:41(6):919–935. 10.1111/j.1365-313X.2005.02341.x [DOI] [PubMed] [Google Scholar]
- Samalova M, Gahurova E, Hejatko J. Expansin-mediated developmental and adaptive responses—a matter of cell wall biomechanics? Quant Plant Biol. 2022:3:e11. 10.1017/qpb.2022.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M, Kirchhelle C, Moore I. Universal methods for transgene induction using the dexamethasone-inducible transcription activation system pOp6/LhGR in Arabidopsis and other plant species. Curr Protoc Plant Biol. 2019:4(2):e20089. 10.1002/cppb.20089 [DOI] [PubMed] [Google Scholar]
- Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jonsson H, Meyerowitz EM. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife. 2014:3:e01967. 10.7554/eLife.01967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005:6(12):242. 10.1186/gb-2005-6-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Montesino R, Bouza-Morcillo L, Marquez J, Ghita M, Duran-Nebreda S, Gomez L, Holdsworth MJ, Basse l, Onate-Sanchez L. A regulatory module controlling GA-mediated endosperm cell expansion is critical for seed germination in Arabidopsis. Mol Plant. 2019:12(1):71–85. 10.1016/j.molp.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M, Trass J. When biochemistry meets mechanics: a systems view of growth control in plants. Curr Opin Plant Biol. 2015:28:137–143. 10.1016/j.pbi.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Scarcelli G, Polacheck WJ, Nia HT, Patel K, Grodzinsky AJ, Kamm RD, Yun SH. Noncontact three-dimensional mapping of intracellular hydromechanical properties by Brillouin microscopy. Nat Methods. 2015:12(12):1132–1134. 10.1038/nmeth.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal F, Wattier C, Rusterucci C, Pelloux J. Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot. 2014:65(18):5125–5160. 10.1093/jxb/eru272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre NBC, Wernerova D, Vittal P, Dubey SM, Medvecka E, Jelinkova A, Petrasek J, Grossmann G, Fendrych M. The AUX1-AFB1-CNGC14 module establishes longitudinal root surface pH profile. bioRxiv. 2022. 10.1101/2022.11.23.517700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004:22(12):1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Takatani S, Verger S, Okamoto T, Takahashi T, Hamant O, Motose H. Microtubule response to tensile stress is curbed by NEK6 to buffer growth variation in the Arabidopsis hypocotyl. Curr Biol. 2020:30(8):1491–1503. 10.1016/j.cub.2020.02.024 [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Higaki T, Umeda M. Actin reorganization triggers rapid cell elongation in roots. Plant Physiol. 2018:178(3):1130–1141. 10.1104/pp.18.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama A, Oka A. ARR1 Directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007:48(2):263–277. 10.1093/pcp/pcl063 [DOI] [PubMed] [Google Scholar]
- Trinh D-C, Alonso-Serra J, Asaoka M, Colin L, Cortes M, Malivert A, Takatani S, Zhao F, Traas J, Trehin C, et al. . How mechanical forces shape plant organs. Curr Biol. 2021:31(3):R143–R159. 10.1016/j.cub.2020.12.001 [DOI] [PubMed] [Google Scholar]
- Vaahtera L, Schulz J, Hamann T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat Plants. 2019:5(9):924–932. 10.1038/s41477-019-0502-0 [DOI] [PubMed] [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. A spatial accommodation by neighbouring cells is required for organ initiation in Arabidopsis. Science. 2014:343(6167):178–183. 10.1126/science.1245871 [DOI] [PubMed] [Google Scholar]
- Vogler H, Caderas D, Mandel T, Kuhlemeier C. Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol Biol. 2003:53(3):267–272. 10.1023/B:PLAN.0000006999.48516.be [DOI] [PubMed] [Google Scholar]
- Voxeur A, Hofte H. Cell wall integrity signalling in plants: “to grow or not to grow that`s the question”. Glycobiology. 2016:26(9):950–960. 10.1093/glycob/cww029 [DOI] [PubMed] [Google Scholar]
- Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, Hong M. Sensitivity-enhanced solid-state NMR detection of expansin's Target in plant cell walls. Proc Natl Acad Sci. 2013:110(41):16444–16449. 10.1073/pnas.1316290110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis L, Refahi Y, Wightman R, Landrein B, Teles J, Huang KC, Meyerowitz EM, Jönsson H. Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. Proc Natl Acad Sci. 2016:113(51):E8238-E8246. 10.1073/pnas.1616768113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Hematy K, Hofte H. Growth control and cell wall signaling in plants. Annu Rev Plant Biol. 2012:63(1):381–407. 10.1146/annurev-arplant-042811-105449 [DOI] [PubMed] [Google Scholar]
- Wu P-J, Kabakova IV, Ruberti JW, Sherwood JM, Dunlop IE, Paterson C, Torok P, Overby DR. Water content, not stiffness, dominates Brillouin spectroscopy measurements in hydrated materials. Nat Methods. 2018:15(8):561–562. 10.1038/s41592-018-0076-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chen H, Huang L, O’Neil RC, Shokhirev MN, Ecker JR. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat Commun. 2018:9(1):1604. 10.1038/s41467-018-03921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Wu MJ, Yan LM, Hu R, Ali I, Gan YB. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLOS One. 2014:9(1):e85208. 10.1371/journal.pone.0085208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, DelRio FW, Ma H, Killaars AR, Basta LP, Kyburz KA, Anseth KS. Spatially patterned matrix elasticity directs stem cell fate. Proc Natl Acad Sci. 2016:113(31):E4439-4445. 10.1073/pnas.1609731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wu Y, Cosgrove DJ. A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 2001:127(1):324–333. 10.1104/pp.127.1.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Hasenstein KH. Distribution of expansins in graviresponding maize roots. Plant Cell Physiol. 2000:41(12):1305–1312. 10.1093/pcp/pcd064 [DOI] [PubMed] [Google Scholar]
- Zhang T, Mahgsoudy-Louyeh S, Tittmann B, Cosgrove DJ. Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose. 2014:21(2):853–862. 10.1007/s10570-013-9996-1 [DOI] [Google Scholar]
- Zhang T, Tang H, Vavylonis D, Cosgrove DJ. Disentangling loosening from softening: insights into primary cell wall structure. Plant J. 2019:100(6):1101–1117. 10.1111/tpj.14519 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu J, Wang X, Durachko DM, Zhang S, Cosgrove DJ. Molecular insights into the complex mechanics of plant epidermal cell walls. Science. 2021:372(6543):706–711. 10.1126/science.abf2824 [DOI] [PubMed] [Google Scholar]
- Zhao H, Brown PH, Schuck P. On the distribution of protein refractive index increments. Biophys J. 2011:100(9):2309–2317. 10.1016/j.bpj.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo YO, Clabaugh Blakley I, Yamburenko MV, Worthen JM, Street HS, Franco-Zorrilla JM, Zhang W, Hill K, Raines T, Solano S, et al. . Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci. 2017:114(29):E5995–E6004. 10.1073/pnas.1620749114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.