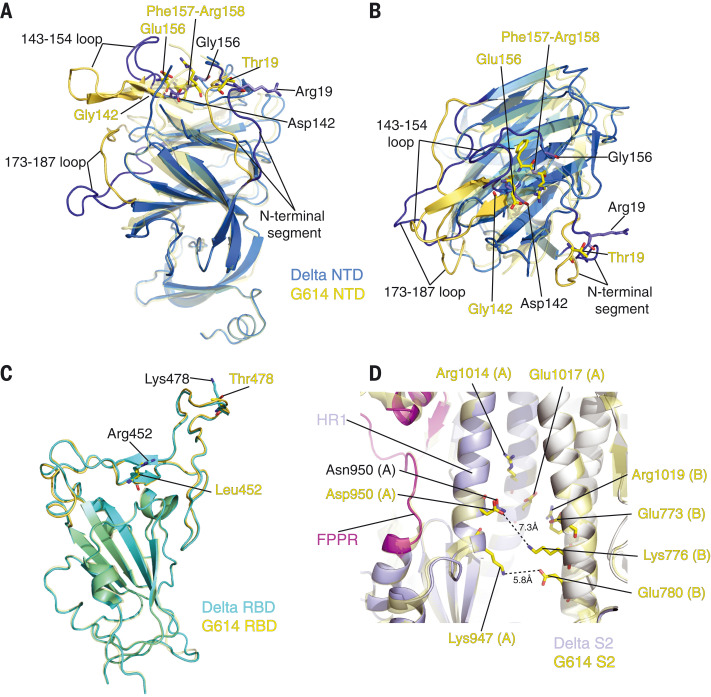
Fig. 4. Structural impact of mutations in Delta S.
(A) Superposition of the NTD structure of the Delta S trimer (blue) with the NTD of the G614 S trimer (PDB ID: 7KRQ) (yellow). Locations of mutations T19R, G142D, E156G, and deletion of F157 and R158 are indicated; these residues are shown in the stick model. The N-terminal segment, as well as loops 143 to 154 and 173 to 187, are rearranged between the two structures and highlighted in darker colors. (B) Top view of panel (A). (C) Superposition of the RBD structure of the Delta S trimer (cyan) with the RBD of the G614 S trimer (yellow). Locations of mutations L452R and T478K are indicated; these residues are shown in the stick model. (D) A close-up view of superposition of the Delta S2 (light blue) with the S2 of the G614 S trimer (yellow) near residue 950. Locations of the D950N mutation and charged residues in the vicinity including Lys947, Arg1014, and Glu1017 from protomer A and Glu773, Lys776, Glu780, and Arg1019 from the protomer B are indicated. All aforementioned residues are shown in the stick model.