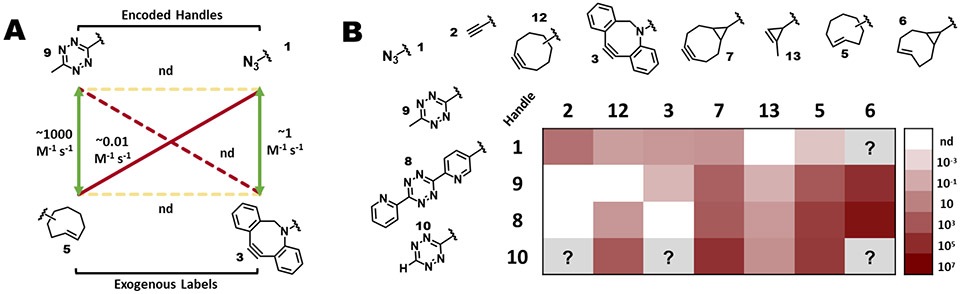
Figure 4.
Identifying chemical orthogonality for reactive groups. A Schematic overview of all possible reactions that can occur during an example dual labeling scenario involving encoded tetrazine 9 and azide 1, and exogenously added TCO 5 and DBCO 3. Depicted are the targeted cognate reactions (green arrows), and the possible off-target reactions between the encoded handles or exogenously added labels (yellow dashed arrows), and between either encoded handle and the non-cognate added label (red arrows; solid arrows indicate observed reactivity and dashed arrows indicate reaction is not detected). For this case, reaction rates (from Karver et al., 2012146) are as indicated for each reaction, with “nd” meaning not detected. As encoded handles that do not react with each other are always selected, and the exogenous labels may be added one after the other, the reactions indicated by the two yellow arrows and one of the red arrows may not be of concern. B Reaction rates of common cycloaddition bioorthogonal handles. Compounds are identified by numbers as indicated. Each panel is color coded (as indicated in the bar to the right) to convey the second order rate constant in M−1 s−1 for the reaction of that pair of compounds. White boxes indicate reaction was not detected (“nd” on the scale bar), and gray boxes “?” indicates that the reaction rate is not published.