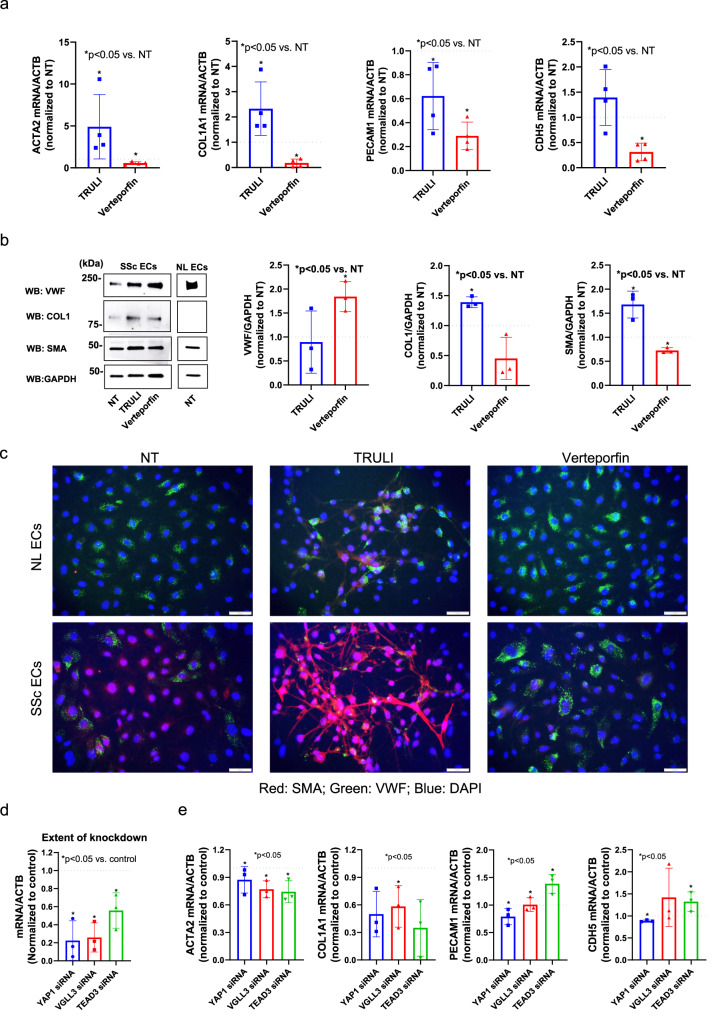
Fig. 5. Hippo pathway regulates endothelial to mesenchymal transition in SSc skin.
a The effect of TRULI (10 μM) and verteporfin (1 μM) on ACTA2, COL1A1, PECAM1, and CDH5 expression in dcSSc endothelial cells. Data normalized to NT. N = 4 patient lines. Data presented as mean +/− SD. Mann–Whitney test was applied. P < 0.05 was designated as statistically significant. P = 0.029 for all the groups that were marked significant. b Western blotting showing the effect of TRULI or verteporfin on VWF, COL1, and SMA in dcSSc endothelial cells. The expression levels of each protein in healthy dermal ECs are shown for comparison. Quantification of samples were from different blots, however, the blots were processed in parallel and the data of each patient line is normalized to its own NT group. N = 3 patient lines. Two-sided unpaired t-test was applied. P < 0.05 was designated as statistically significant. P = 0.0095 for VWF-verteporfin, p = 0.0016 for COL1-TRULI, p = 0.014 for SMA-TRULI, p = 0.001 for SMA-vertepofrin. NL normal. c Immunofluorescence showing TRULI enhanced the mesenchymal phenotype while verteporfin promoted the endothelial phenotype in dcSSc endothelial cells, while in healthy ECs, TRULI induced EndoMT to a lesser extent, while verteporfin had minimal effect. Images shown are representative of N = 3 patient lines. Scale bar = 50 μm. d The extent of knockdown of YAP1, VGLL3, or TEAD3 in dcSSc endothelial cells. N = 3 patient lines. Data normalized to control and presented as mean +/− SD. Two-sided unpaired t-test was applied. P < 0.05 was designated as statistically significant. P = 0.0036 for YAP1 siRNA, p = 0.0013 for VGLL3 siRNA, p = 0.017 for TEAD3 siRNA. e Knockdown of genes involved in the Hippo pathway blocked the EndoMT phenotype in dcSSc endothelial cells. Two-sided unpaired t-test was applied. P < 0.05 was designated as statistically significant. Data presented as mean +/− SD. N = 3 patient lines. ACTA2: p = 0.012 for VGLL3 siRNA, p = 0.02 for TEAD3 siRNA; COL1A1: p = 0.025 for YAP1 siRNA, p = 0.033 for VGLL3 siRNA, p = 0.022 for TEAD3 siRNA; PECAM1: p = 0.018 for TEAD3 siRNA; CDH5: p = 0.0007 for YAP1 siRNA. Source data is provided for this figure.