Figure 3.
Relieving Par6 autoinhibition is the rate-limiting step in Par complex assembly
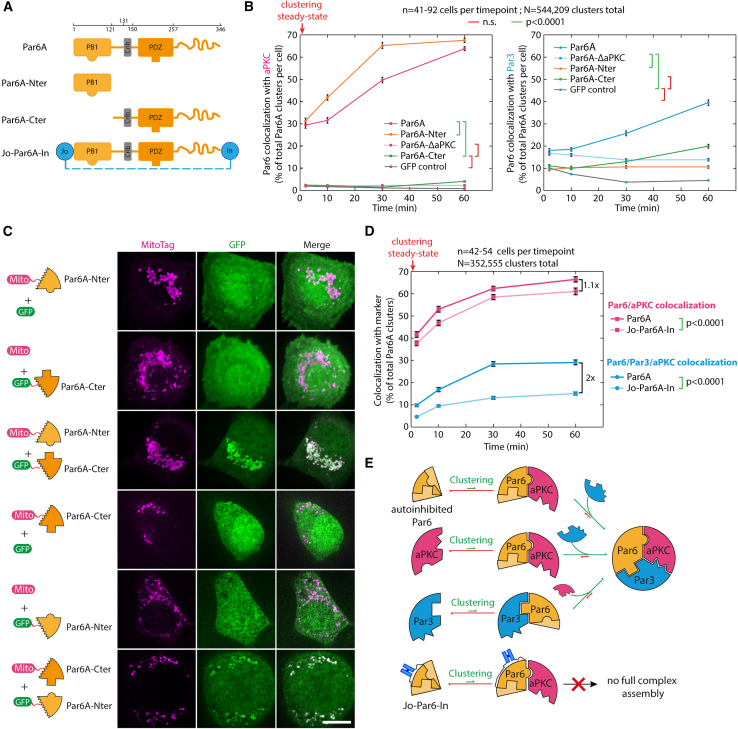
(A) Par6A fragments used in this study. The N-terminal PB1 domain of Par6A binds to aPKC, while the C-terminal PDZ domain binds to Par3. The Jo domain, fused to the N terminus, covalently binds to the In domain, fused to the C terminus.32
(B) Mean (±SEM) percentage of colocalization between GFP-Par6A or Par6A mutant clusters and aPKC (left) or Par3 (right). Statistics: right: ANOVA to test interaction with time: Par6A: p < 0.0001; Par6ACter: p < 0.0001; Par6ANter: n.s.; Par6AΔaPKC: p < 0.01. left: ANOVA2 using construct and time point as variables (p value of construct effect indicated). Par6A/GFP curves are the same as in Figure 2, reproduced here for convenience. See Figure S7B for representative images.
(C) Par6ANter binds Par6ACter. 3T3 cells expressing cytosolic GFP-Par6ACter, and Par6ANter tethered to the mitochondria, or vice versa, were imaged by SDCM. Maximum intensity z-projection (MIP) is shown. Par6ANter recruits Par6ACter to mitochondria, and vice versa, suggesting that Par6A folds on itself (see also Figure S7C for further controls).
(D) GFP-fused Par6A, or Jo-Par6-In, was clustered in 3T3 cells, and the recruitment of endogenous aPKC and Par3 was measured over time (mean ± SEM percentage of dual colocalization between GFP-Par6A clusters and aPKC [magenta] and triple colocalization between Par6A, aPKC, and Par3 [blue]). See Figure S7F for representative images. Statistics: ANOVA2 using construct and time point as variables (p value of construct effect indicated). Note that the Jo-Par6A-In profoundly inhibits assembly of the tripartite Par complex.
(E) Preferred route for assembly of the core Par complex as a function of the clustered subunit (see also STAR Methods).
Scale bars: 10 μm in (C).