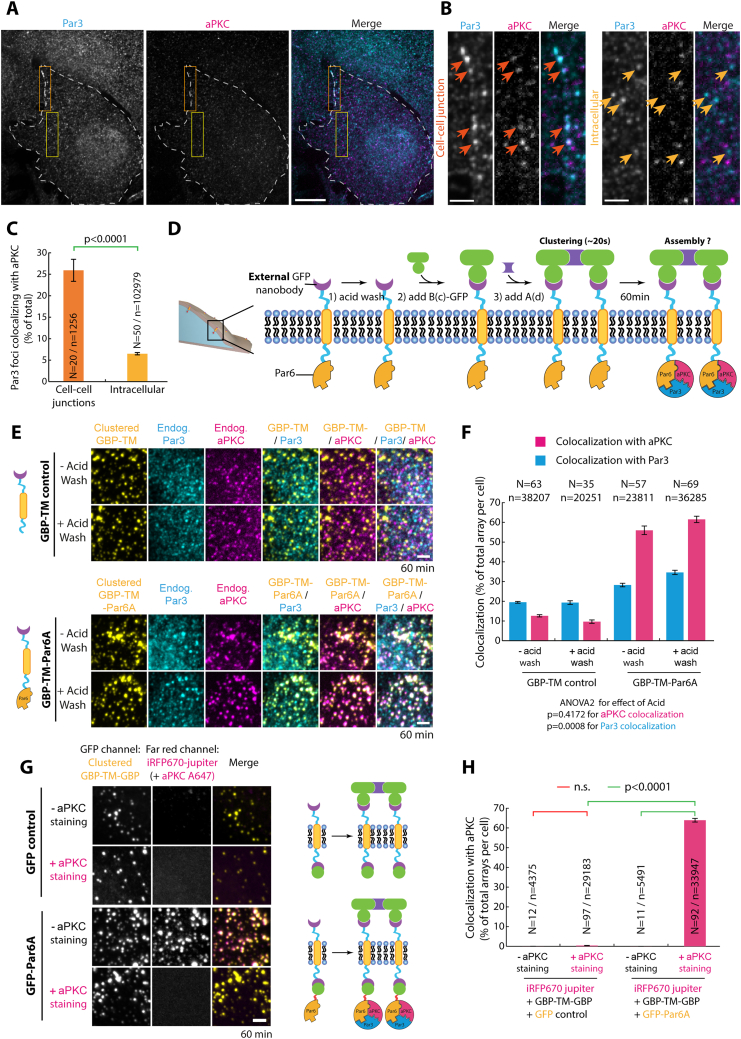
Figure S3.
Determination of physiological Par complex assembly at cell-cell junctions in 3T3 cells and control experiments of the bicistronic clustering method, related to Figure 2
(A) 3T3 cells in interphase immunostained for endogenous Par3 and aPKC and imaged by SDCM. Images correspond to MIPs of 7 planes (Δz = 1.4 μm total). Dashed white line: cell contours.
(B) High magnification of the image presented in (A) in a cell-cell junction region (left, corresponds to orange rectangle in (A) or in an intracellular region (right, corresponds to yellow rectangle in A). Arrows indicate Par3 clusters colocalizing with aPKC at cell-cell junctions in contrast to the rest of the cell.
(C) Mean ± SEM percentage of colocalization between Par3 clusters and aPKC per region (see also STAR Methods for details about automated 3D object-based colocalization). Statistics: unpaired Student’s t test (N: regions of interest analyzed; n: total number of Par3 spots detected).
(D–F) The acid wash treatment ensuring that the external GBP is not bound to GFP does not affect the colocalization between Par6A and aPKC/Par3 used in this study.
(D) Principle of the experiment: Cells are treated with a quick acid wash prior to GBP-TM-GBP clustering to avoid extracellular GBP from being saturated with GFP fusion proteins in the culture medium (released upon cell death, for instance).
(E) Cells processed as in (D) were immunostained for endogenous aPKC and Par3 after 60 min of clustering. Imaging was performed by SDCM (MIPs: Δz = 3.8–4.6 μm total).
(F) Mean ± SEM percentage of the 3D colocalization between GFP-Par6A (or GFP control) clusters and aPKC or Par3 per cell. Effect of the acid wash was tested using an ANOVA2 test (respective p value indicated).
(G and H) The weak iRFP670-Jupiter signal in the far-red channel does not affect the accuracy of the detection the strong signal of the Alexa-647 aPKC immunostaining.
(G) Cluster formed at the surface of 3T3 cells expressing iRFP670-Jupiter, GBP-TM-GBP, and GFP-Par6A (or GFP control) were immunostained (or not) for endogenous aPKC using Alexa647-coupled secondary antibodies and imaged by SDCM (MIPs; Δz = 3.4–4.6 μm total). Dynamic range was set to be identical between images.
(H) Mean ± SEM percentage of the 3D colocalization between Par3 clusters and aPKC per cell. Statistics: Kruskal-Wallis test followed by a Dunn post hoc test (p value of respective tests indicated).
In (F) and (H), N corresponds to the number of cells analyzed per condition, and n corresponds to the total number of GFP-positive arrays detected per condition.
All images in this figure were processed with a wavelet “a trous” filter (see STAR Methods). Scale bars: 10 μm in (A) and 2 μm in (B), (E), and (G).