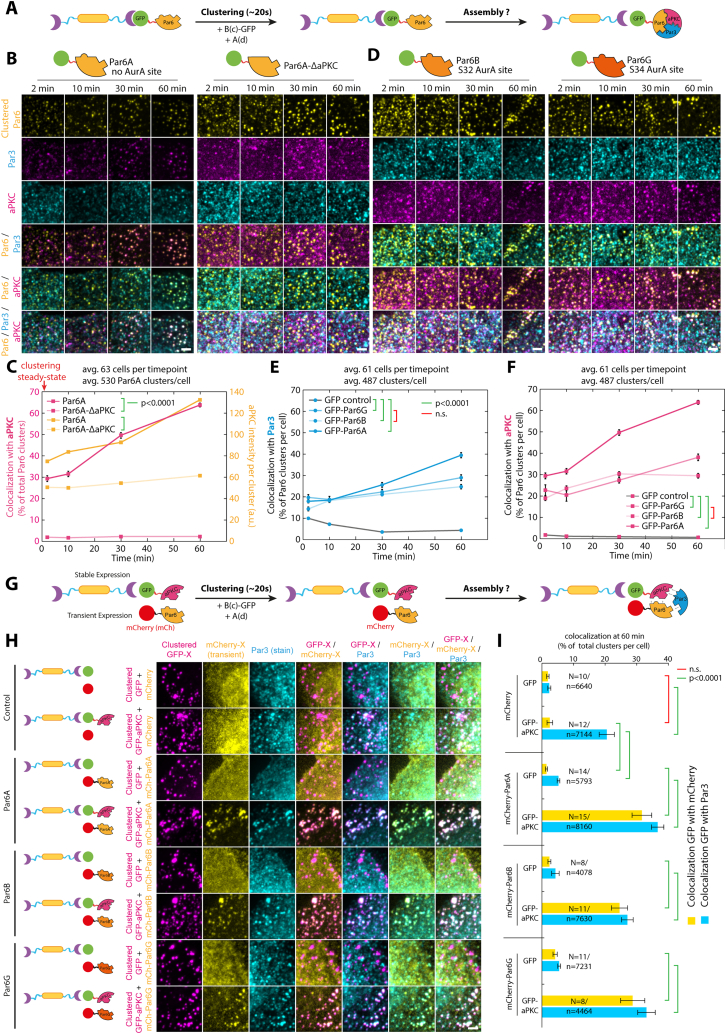
Figure S4.
Characterization of the ability of Par6 isoforms to support Par complex assembly, related to Figure 2
(A) Principle of the experiment: 3T3 cells stably co-expressing GBP-TM-GBP and GFP-fused Par6 variants were incubated with B(c)GFP then A(d) to induce rapid clustering, and the recruitment of endogenous aPKC and Par3 was monitored by immunofluorescence.
(B) SDCM images of cells expressing indicated Par6 mutant were treated as presented in (A) and immunostained for Par3 and aPKC.
(C) Mean ± SEM percentage of colocalization between GFP-Par6A (or GFP-Par6AΔaPKC) clusters and aPKC over time, plotted at the same time as the mean ± SEM intensity of aPKC per cluster over time.
(D) SDCM images of cells expressing Par6 isoforms, treated as in (A) then stained for Par3 and aPKC.
(E and F) Mean ± SEM percentage of colocalization between clusters of indicated Par6 isoform and Par3 (E) or aPKC (F). Statistics: ANOVA2 using construct and time point as variables followed by Tukey post hoc test (p value of each test indicated).
(G) Principle of the experiment: 3T3 cells stably expressing GBP-TM-GBP and GFP-fused aPKC and transiently transfected with mCherry-Par6 isoforms were incubated with B(c)GFP then A(d) to induce rapid clustering.
(H) Cells expressing indicated Par6 isoform (or controls) were treated as presented in (G), and the recruitment of exogenous Par6 and endogenous Par3 to aPKC clusters was monitored by immunofluorescence.
(I) Mean ± SEM percentage of colocalization between clusters of aPKC-GFP and indicated mCherry-fused Par6 isoform and Par3. Statistics: ANOVA1 using construct and time point as variables followed by Tukey post hoc test (p value of each test indicated).
All images in this figure were processed with a wavelet a trous filter (see STAR Methods). Scale bars, 2 μm.
See also Table 2 for definition of the constructs used in this figure.