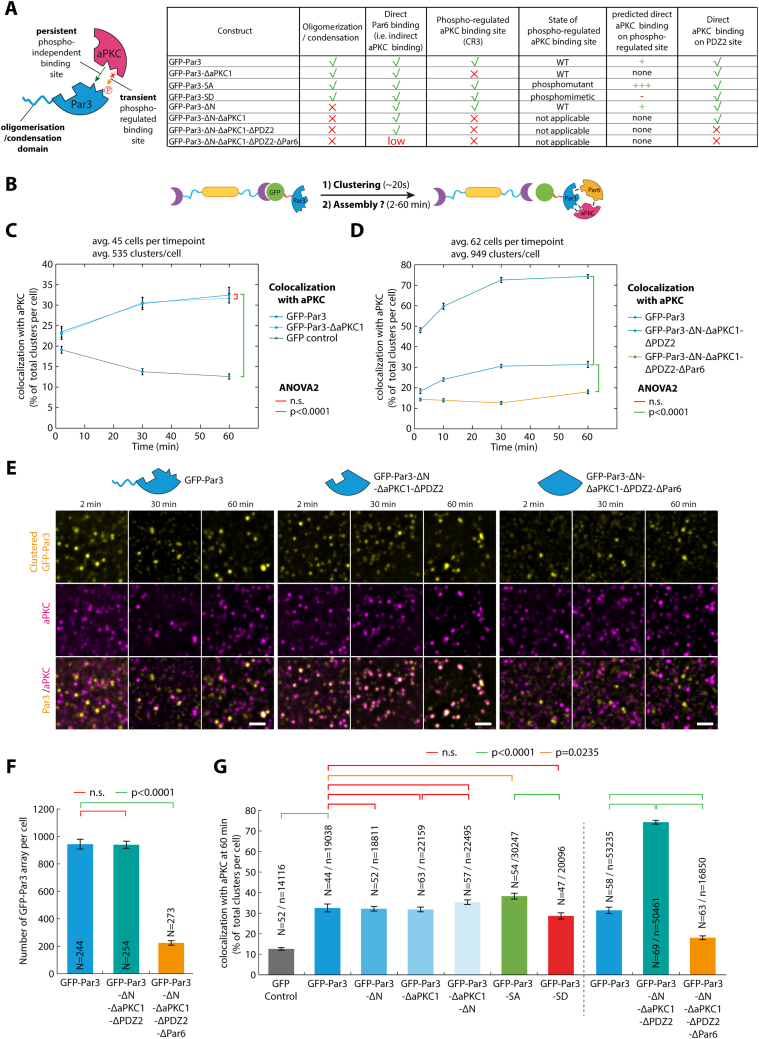
Figure S6.
Characterization of the effect of Par3 mutants on Par complex assembly, related to Figure 2
(A) Description of the constructs used in this figure and their predicted effects on oligomerization, aPKC binding, and Par6 binding. Par3 has two binding sites for aPKC. One interaction takes place between the kinase domain of aPKC and the aPKC phosphorylation motif on Par3 (also known as CR3 domain). This interaction is phospho-regulated and abolished upon phosphorylation. A second site on Par3, the PDZ2 domain, binds to the PBM domain of aPKC in a phosphorylation-independent way.31 In addition, Par3 oligomerization/condensation is known to occur via the N-terminal domain.43
(B) Principle of the experiment: 3T3 cells stably expressing GBP-TM-GBP and GFP-fused Par3 (or mutant thereof), were incubated with B(c)GFP then A(d) to induce rapid clustering. Then, the assembly of the endogenous Par complex was followed over time by aPKC immunofluorescence and automated quantification.
(C and D) Mean ± SEM percentage of colocalization between clusters of indicated GFP-Par3 construct and aPKC as a function of time after cluster formation (time point 0). Statistics: ANOVA2 using time and construct as variables; p value for effect of the construct indicated.
(E) Cells expressing indicated Par3 mutant were treated as in (B), and the recruitment of endogenous aPKC was monitored by immunofluorescence. Images were processed with a wavelet a trous filter. These images correspond to the samples quantified in (D). Scale bars, 2 μm.
(F) Mean ± SEM of the number of GFP-Par3 array per cell in the samples presented in (D) and (E). Statistics: Kruskal-Wallis test followed by Dunn’s post hoc test (p values figured for each test, n indicates the number of cells averaged). Note that Par3-ΔN-ΔaPKC1-ΔPDZ2 has a propensity to make fewer clusters than Par3 and Par3-ΔN-ΔaPKC1.
(G) Mean ± SEM of the aPKC/Par3 colocalization 60 min post clustering for the indicated construct. Statistics: one-way ANOVA followed by Dunn’s post hoc test (p values figured for each test, n indicates the number of cells averaged).
See also Table 2 for definition of the constructs used in this figure.