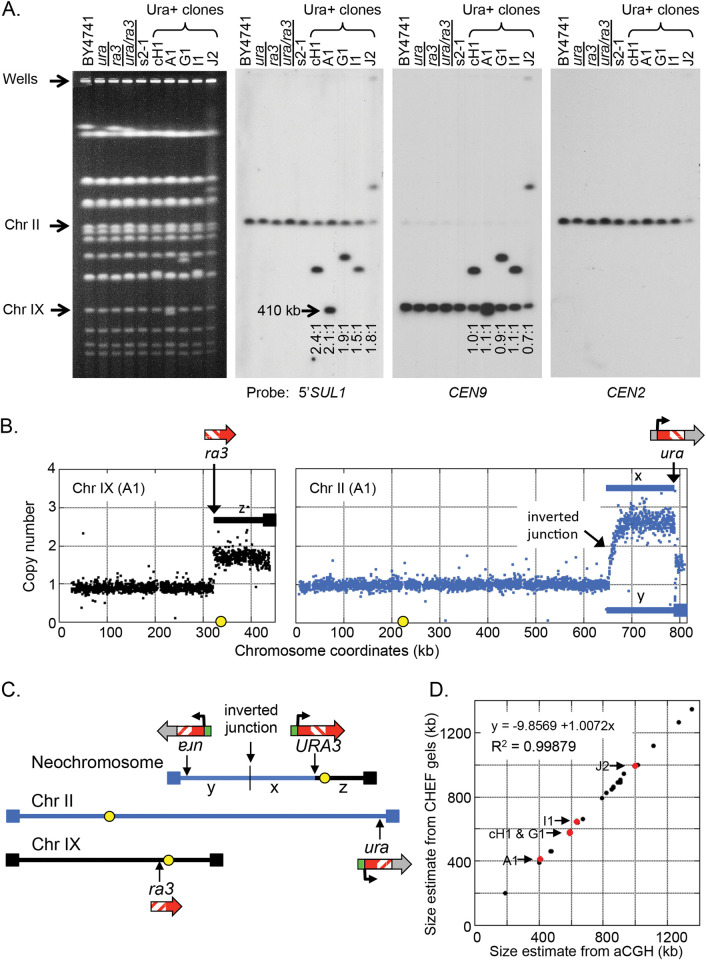
Fig 3. Physical characterization of chromosomes from Ura+ clones.
(A) Chromosomes from a wild type strain, the haploid ura and ra3 strains, the heterozygous diploid strain, split-ura3 strain, and five Ura+ clones were analyzed by CHEF gel electrophoresis and Southern blot hybridization. The ethidium bromide stained gel and the Southern hybridizations reveal that the Ura+ clones contain an additional single unique chromosome that hybridizes to the centromere of chromosome IX and also to the 5’SUL1 probe. The ratio of 5’SUL1 hybridization for each of the five Ura+ strains shows a ~2:1 hybridization ratio for the neochromosome relative to the native chromosome II (measured by probe signal). (B) Array comparative genome hybridization of clone A1 vs. the parent split-ura3 strain indicates that the right telomeric segment of chromosome IX (labeled z) and the ~20 kb at the right telomere of chromosome II (as part of the fragment labeled y) are present at a copy number of ~2. A larger subtelomeric segment of chromosome II (labeled x) is present at a copy number of ~3. The sites of the ura and ra3 insertions are indicated by arrows. (C) The proposed structure of the inverted neochromosome that contains the three amplified segments x, y, and z is illustrated above the two non-rearranged chromosomes. (D) The sizes of each of the five inverted neochromosomes (red dots) deduced from CHEF gels correspond to their predicted sizes based on aCGH data. Size estimates of non-inverted Ura+ chromosomes are indicated by black dots.