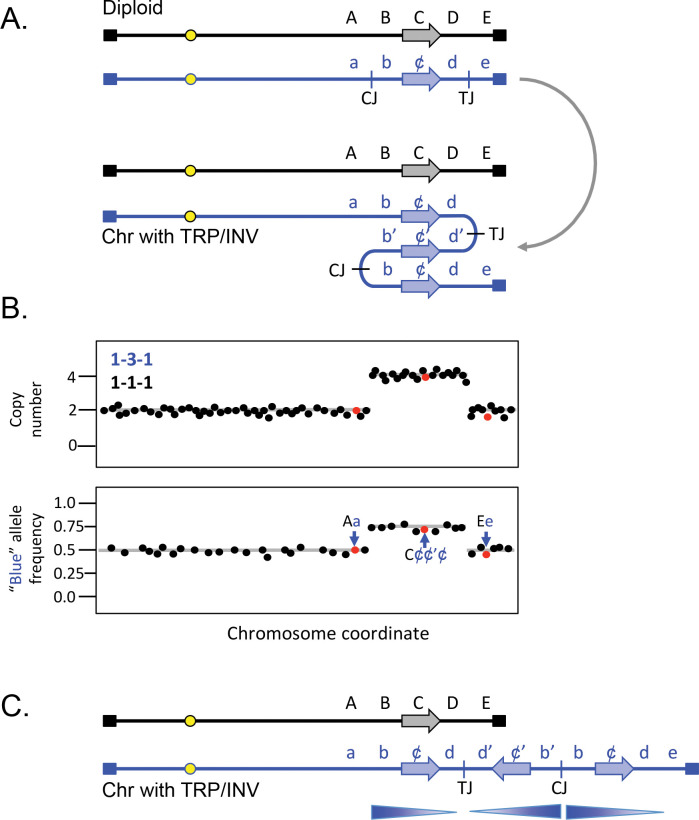
Fig 2. Inverted triplication.
(A) A generic example of an inverted triplication in a diploid, affecting the blue chromosome, with SNPs indicated in upper and lower case letters (lower case “c” being depicted as ¢ for clarity). The horizontal arrow represents a potential coding sequence. CJ and TJ refer to the potential centers of the inversion junctions (centromere-proximal and telomere-proximal junctions) identified after the inversion and triplication of the segment containing the b, c, and d SNPs. The derived chromosome is shown folded back on itself to emphasize the triplication and the inverted center copy. (B) Top; expected copy number results (using either aCGH or read depth) of the diploid after triplication of the b–d region. Bottom; allele frequencies for SNPs unique to the blue chromosome. (C) Linear representation of the 2 homologues after triplication affecting the blue chromosome. Arrows indicate the orientation of the 3 segments involved in the triplication. Notice that the right end of the chromosome remains intact. aCGH, array comparative genome hybridization.