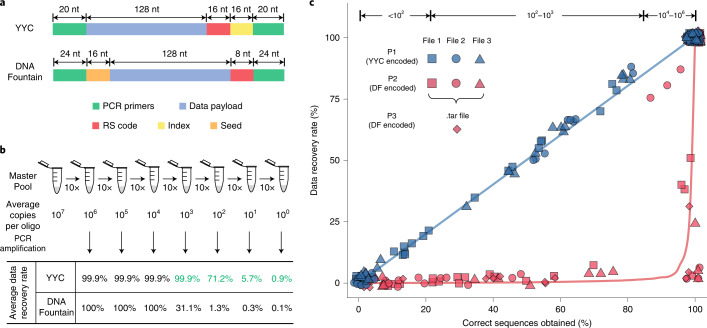
Fig. 3. Experimental validation of in vitro binary data storage using the YYC and DNA Fountain coding strategies.
a, The sequence design of the 200 nt oligo generated by the YYC and DNA Fountain algorithms for in vitro data storage. b, The serial dilution experiment of the synthesized oligo pool. The average copy number of each oligo sequence is calculated accordingly to the original oligo pool. The average data recovery rates are calculated based on the sequencing result of the PCR products of the diluted samples (the data recovery rates of YYC samples with low molecule copy number (≤103) is labeled with green color; the DNA molecule copy number for each sample after the PCR amplification exceeds 108). c, Analysis of the YYC and DNA Fountain (abbreviated as DF) algorithms by sequencing of corresponding diluted samples and calculation of the data recovery rate of each file encoded by YYC and DNA Fountain at the corresponding oligo copy number.