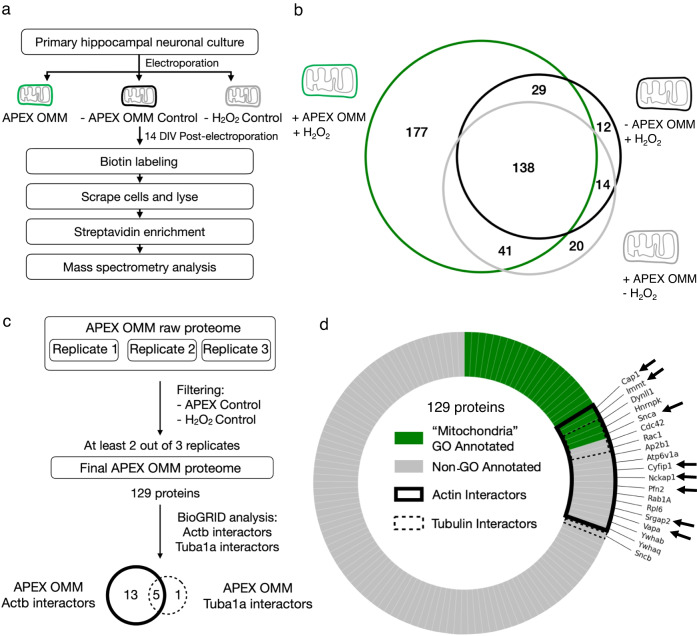
Fig. 2. Identifying actin-interacting proteins on the outer mitochondrial membrane.
a Experimental workflow for APEX-OMM proteome labeling in primary hippocampal neuronal cultures for mass spectrometry analysis. b Representative biological replicate (of c, d) showing the OMM proteome yield (green) after subtracting the proteins measured in Controls—in the absence of APEX-OMM (black) and the absence of hydrogen peroxide (gray). The proteins detected in Controls are endogenous biotinylated proteins (pyruvate carboxylase, 3-methylcrotonyl CoA carboxylase, propionyl CoA carboxylase, and acetyl CoA carboxylase) and non-specific proteins binding to streptavidin beads during enrichment. As we followed a stringent criterion of excluding the proteins found in Controls from experimental samples, irrespective of their intensities (see Methods), there might be an overestimation of proteins in the two Control samples. c Flowchart of the mass spectrometry proteome analysis of three biological replicates, following filtering for Controls as in (b), yielding 129 proteins from two out of three biological replicates and identifying actin (Actb) and tubulin (Tuba1a) interactors on the OMM using BioGRID. d 129 proteins of the OMM proteome analyzed for gene ontology (GO) annotation for the term “Mitochondria” (“Mitochondria GO Annotated, green), and proteins not GO annotated as “Mitochondria” (Non-GO Annotated, gray). 18 proteins identified in BioGRID as actin interactors (Actin Interactors, black line), 6 proteins identified in BioGRID as tubulin interactors (Tubulin Interactors, dotted black line), and the 8 actin-interacting OMM proteins selected for the next round of screening (black arrows). n: 3 biological replicates, 3 animals.