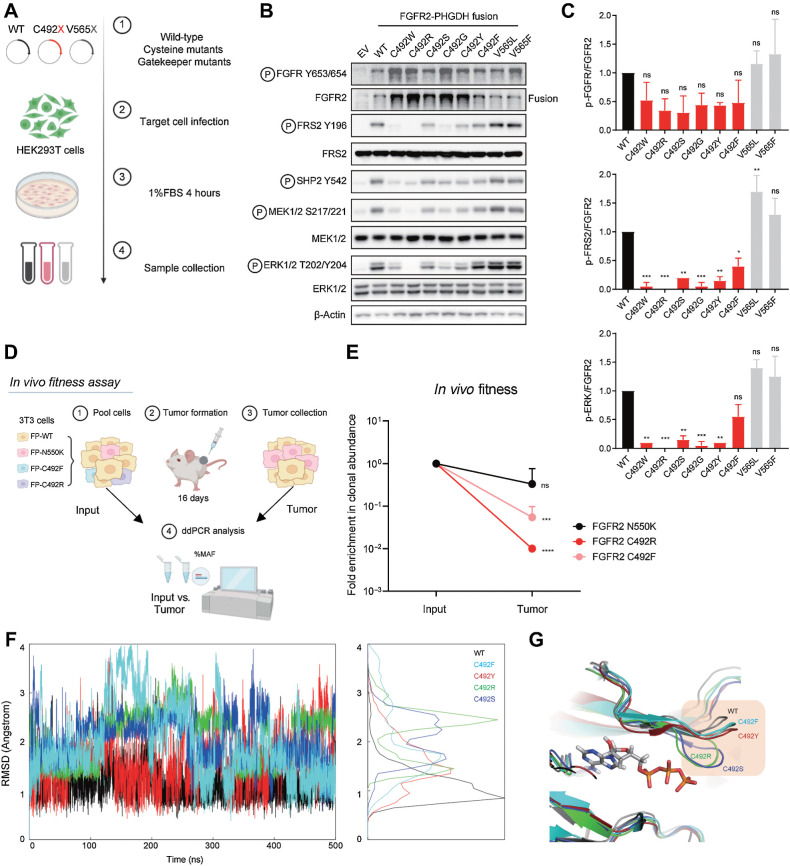
Figure 4.
Mutation of FGFR2 C492 compromises FGFR2 signaling. A–C, Signaling analysis of FGFR2 fusion alleles harboring mutations of C492. A, Schematic view of signaling studies. 293T cells were engineered by retroviral transduction to stably express empty vector (EV) or the FGFR2–PHGDH fusion harboring a WT kinase domain, each mutation generated by single nucleotide changes at FGFR2 codon 492, or mutations at the gatekeeper (V565) residue. Immunoblot analysis of signaling proteins (B) and quantification (C). Quantifications were generated from two independent experiments. Data are shown as mean ± SD. Statistical comparison of WT vs. C492 variants by one-way ANOVA (C). D and E, Clonal fitness of cysteine mutations. D, Schematic view of in vivo fitness assay. NIH-3T3 cells engineered with the indicated mutants were pooled and injected subcutaneously into NSG mice. After 16 days, tumors were harvested and processed for ddPCR analysis. E, Clonal abundance of indicated mutants in vivo. Each point represents 3 replicates. Data are shown as mean ± SD. Statistical analysis of input versus tumor by unpaired t test. F–G, Molecular dynamics simulations of select FGFR2 C492 mutations. F, Fluctuations of the G-loop measured by root mean square deviation (RMSD). G, Representative structures from the simulation trajectory. (A and D, Created with BioRender.com. F, Created with R. G, Created with PyMOL.)