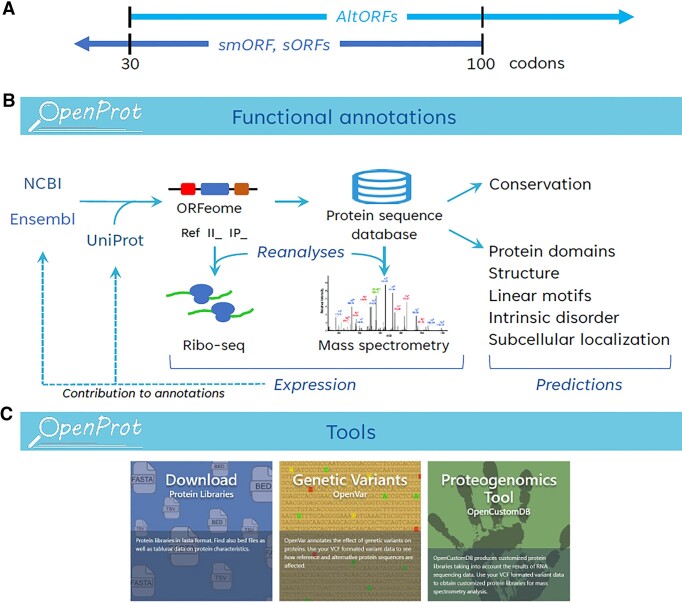
Figure 1.
(A) AltORFs are defined as ORFs with a minimum size of 30 codons (including the stop codon) currently unannotated in RefSeq and Ensembl, AltORFs can be larger than 100 codons. The longest AltORFs in human has 13 226 codons (II_4421741). Small ORFs (smORFs or sORFs) are typically 100 codons or less in length. (B) OpenProt functional annotations. Interrogation of Ensembl and RefSeq transcripts results in the identification of annotated ORFs (or RefORFs) and unannotated ORFs (or AltORFs). The corresponding proteins are classified as RefProts, novel isoforms (accession II_) or AltProt (accession IP_). Reanalyses of ribo-seq and mass spectrometry-based proteomics datasets with the OpenProt databases provide expression evidence and contribute to the addition of AltProts and novel isoforms into standard databases (dotted arrows; Table 1). Functional predictions help identify AltProts and novel isoforms with potential biological activity. (C) OpenProt tools. Protein sequence databases are available for download. OpenVar is a genomic variant annotator able to handle multiple ORFs in a single transcript. OpenCustomDB allows users to build customized protein databases from RNAseq data, integrating genetic variants in RefProts, AltProts and novel isoforms.