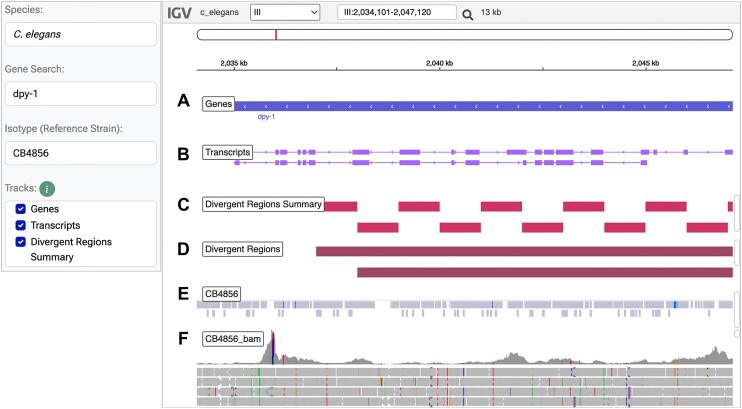
Figure 1.
The CaeNDR genome browser. The user specifies the Species in the setup panel to the left. The optional Gene Search field will direct the browser to that gene in the genome (A) and display transcripts in the track below (B). The Divergent Regions Summary track shows regions of the genome that are classified as hyper-divergent relative to the reference genome in any isotype reference strain within the species (C). The Divergent Regions track displays regions that are hyper-divergent relative to the reference genome for individual isotype reference strains, each track feature can be clicked to reveal the isotype reference strain the feature belongs to (D). The Isotype field in the setup window on the left is optional and allows users to add VCF and BAM tracks to the browser. The VCF track shows species-wide SNV and short indels in gray, with the variants specific to the isotype reference strain in blue (E). The BAM track shows read level data from which the variants were called. The top of the track shows read depth and the bottom shows the paired-end reads (F).