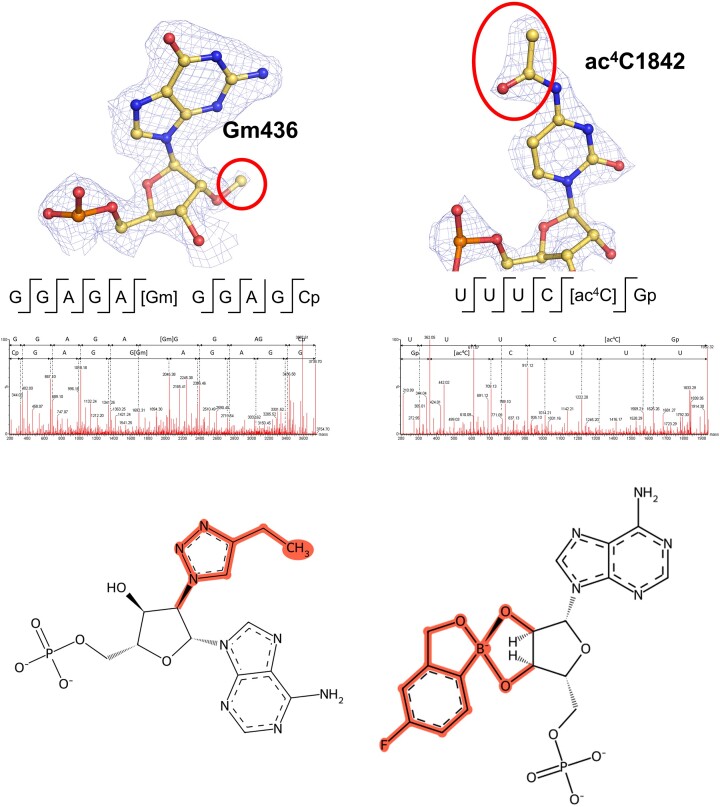
Figure 1.
Illustration of the key new features in MODOMICS. Top panel: Examples of modifications sites identified in rRNAs using high-resolution cryo-EM and fragment-based MS analysis of 80S ribosomes. The modified ribonucleotides are shown in the context of the Coulomb density map displayed as blue mesh, with modifications indicated by red ellipsoids. Below, the validation by mass spectrometry using enzyme-induced RNA fragment analysis is illustrated. Bottom panel: 2D schemes of ribonucleotide residues imported to MODOMICS: 2′-deoxy-2′-(4-ethyl-1H-1,2,3-triazol-1-yl)adenosine 5′-(dihydrogen phosphate), left; [(6-amino-9H-purin-9-yl)-[5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole]-4′yl]methyl dihydrogen phosphate (ANZ), right. Chemical modifications are highlighted in red.