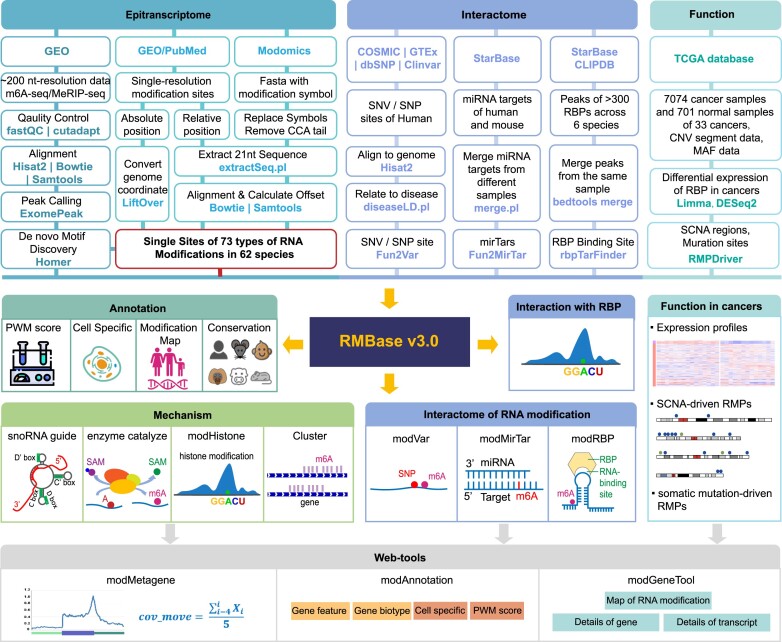
Figure 1.
The workflow of RMBase v3.0. RMBase v3.0 serves as a comprehensive resource for deciphering RNA modification maps, biogenesis mechanisms, functions, interactomes, evolutionary conservation, and disease variations. It explores RNA modification landscapes across 62 species by de novo identifying m6A sites from ∼200 nt resolution high-throughput sequencing data and integrating modifications with single nucleotide precision from Modomics and publications (top left). RMBase v3.0 assigns scores to each RNA modification site using PWMs, enriches annotations with genomic information, cell lines and experimental datasets, and unveils their evolutionary conservation in different mammals. The platform investigates RNA modification biogenesis mechanisms at both genome and transcriptome levels (middle left). It delves into the interactome of RNA modifications with RBPs, miRNA targets, SNPs, and SNVs, while elucidating the biological roles of RMPs in tumors through analyzing RNA-seq from normal and tumor samples (top right and middle right). Based on the above-mentioned data, RMBase v3.0 also provides three web-based tools (bottom) for users to perform customized analyses.