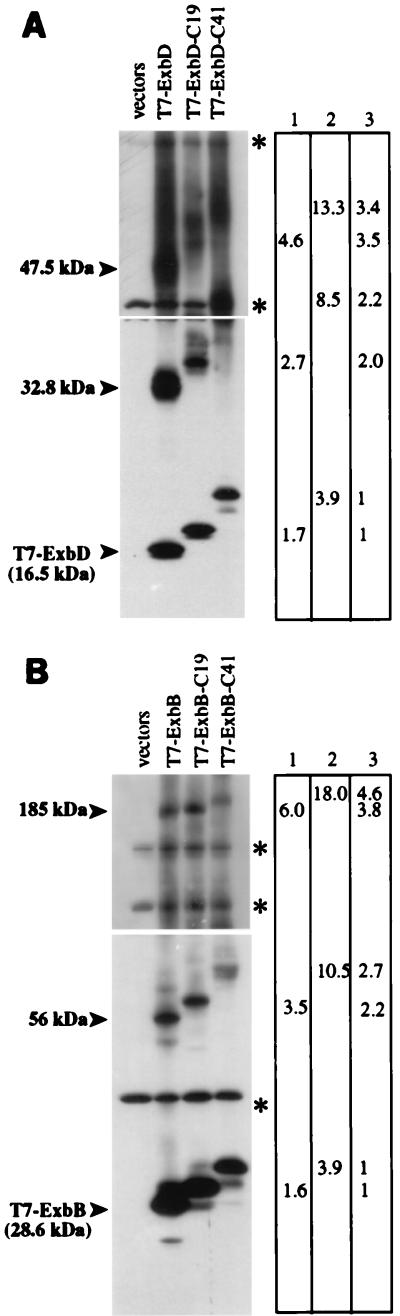
FIG. 6.
In vivo cross-linking of T7-ExbB or T7-ExbD size variants. (A) Strain KP1269 (ΔexbBD) carrying plasmids pKP361 (ExbB) and pKP323 (T7-ExbD), pKP375 (T7-ExbD-C19), or pKP377 (T7-ExbD-C41). (B) Strain KP1269 (ΔexbBD) carrying plasmids pKP360 (ExbD) and pKP339 (T7-ExbB), pKP353 (T7-ExbB-C19), or pKP376 (T7-ExbB-C41). Cells were cross-linked in vivo by using standard formaldehyde as described in Materials and Methods. Samples representing equal cell numbers were electrophoresed on SDS–7 to 20% polyacrylamide gradient gels, and complexes containing T7 epitope-tagged proteins were detected by immunoblot analysis with an anti-T7 epitope tag monoclonal antibody. The molecular masses of complexes were measured relative to those of unstained protein standards (BenchMark; Gibco BRL). Columns: 1, measured differences in molecular mass between T7-Exb-C19 complexes and T7-Exb complexes (in kilodaltons); 2, measured differences in molecular mass between T7-Exb-C41 complexes and T7-Exb complexes (in kilodaltons); 3, factor increase in molecular mass. Longer exposures (top) were necessary for detection of the higher-molecular-mass complexes; shorter exposures (bottom) were necessary for accurate relative mobility measurement of lower-molecular-mass complexes. ∗, cross-reactive band.