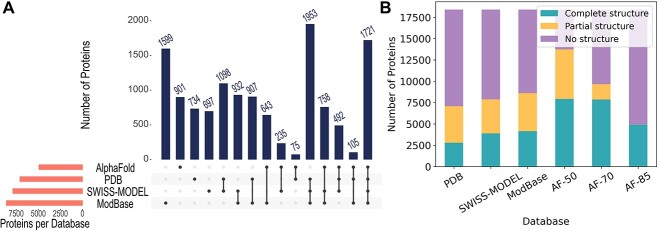
Figure 2.
Structural coverage of proteins in the human reference proteome by databases. (A) The number of proteins modeled by PDB, SWISS-MODEL, ModBase, AF and their intersections are visualized. AF models with 85% of their residues predicted with ≥70% pLDDT score are used. (B) The number of proteins with no structure, partial structure, and complete structure. AF models with 85% (AF-85), 70% (AF-70) and 50% (AF-50) of their residues predicted with ≥70% pLDDT score are used for this demonstration. Partial structure denotes structure coverage <80% for PDB and homology models. For AF, it denotes an average accuracy of <80%. Similarly, complete structure means ≥80% coverage or average accuracy. Although it’s not visible, AF-85 has 13 models with partial structures.