Figure 1.
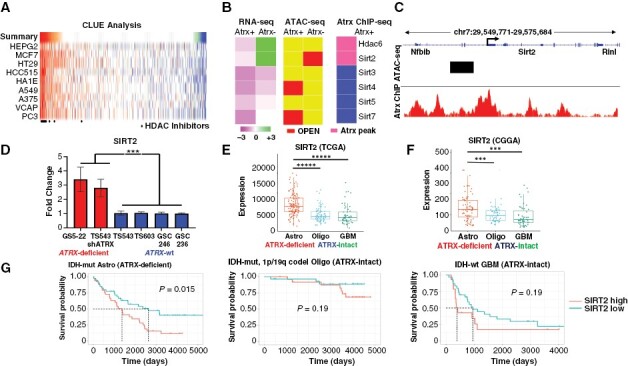
SIRT2 is upregulated in ATRX-deficient glioma. (A) Heatmap of CMAP analysis of Atrx-deficient transcriptional changes in mNPCs demonstrating strong negative correlations with several HDAC inhibitors. (B) Heatmaps showing transcript levels (RNA-seq), proximity to regions of increased chromatin accessibility (ATAC-seq; OPEN), and proximity to ATRX binding sites (Atrx ChIP-seq), of various HDACs in mNPCs (Atrx status shown). (C) Schematic of the Sirt2 locus showing associated Atrx binding peaks (Atrx ChIP) and region of increased chromatin accessibility in Atrx− mNPCs (ATAC-seq). (D) RT-PCR analysis showing increased SIRT2 expression in ATRX-mutant (GS5-22), and ATRX-knockdown (TS543-shATRX) GSCs, compared to ATRX-wildtype (TS543, TS603, GSC 246, and GSC 236) GSCs. Expression analysis in TCGA (E) and CGGA (F) datasets revealed increased SIRT2 levels ATRX-deficient gliomas relative to ATRX-intact counterparts. (G) Kaplan–Meier analysis showing that high SIRT2 expression is associated with poor prognosis in ATRX-deficient gliomas but not in ATRX-intact counterparts. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
