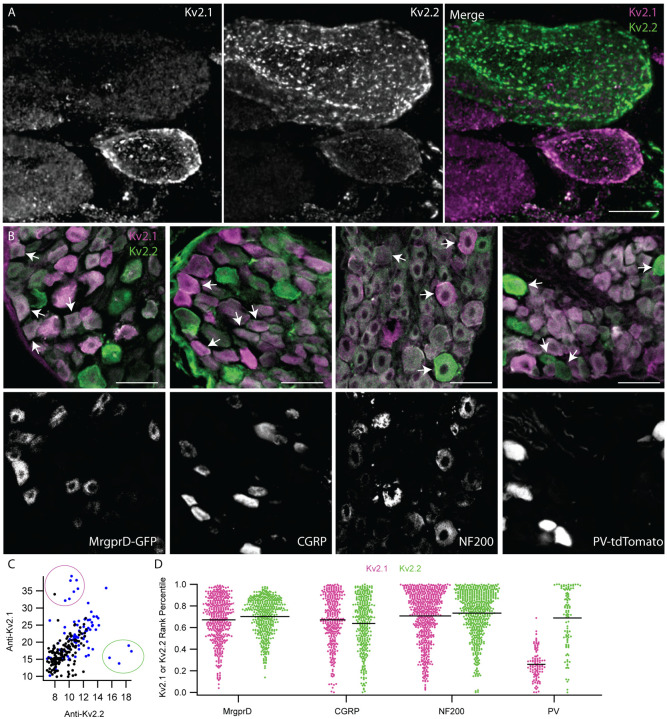
Figure 9.
Anti-Kv2 immunofluorescence intensities are non-uniform across DRG neuron subtypes. A, Exemplar z-projection of enrichment of anti-Kv2.1 (magenta) or anti-Kv2.2 (green) immunofluorescence in neighboring neurons. DRG section is from a 10 week old male mouse. Scale bar is 10 μm. B, Top images show anti-Kv2.1 (magenta) and anti-Kv2.2 (green) immunofluorescence in DRG sections where subpopulation specific markers were used to identify, from left to right, non-peptidergic nociceptors, peptidergic nociceptors, myelinated neurons and proprioceptors. Fluorescence from specific markers is shown in bottom panels. Arrows indicate four exemplar neurons that have clear positivity for each subpopulation identified by fluorescence in lower panels. CGRP and NF200 subpopulations were identified using anti-CGRP and anti-NF200 antibodies while MrgprD-GFP and PV-tdTomato subpopulations were from transgenic mouse lines. Scale bars are 50 μm. C, Scatter plot of anti-Kv2.1 and anti-Kv2.2 immunofluorescence of individual neuron profiles. Each point represents one profile. Magenta circle highlights the subpopulation of profiles that have high anti-Kv2.1 but low anti-Kv2.2 immunofluorescence while the green circle highlights the subpopulation of profiles that have high anti-Kv2.2 but low anti-Kv2.1 immunofluorescence. Blue points represent myelinated DRG neuron profiles identified by NF200 immunofluorescence. D, Ranked anti-Kv2.1 immunofluorescence (magenta points) or ranked anti-Kv2.2 immunofluorescence (green points) of individual profiles from subpopulations shown in B. Only profiles that were positive for each marker are shown. Each point represents one profile. MrgprD population N = 4 mice, CGRP population N = 3 mice, NF200 population N = 3 mice and PV population N = 2 mice. Detailed information on each mouse used can be found in table 1.