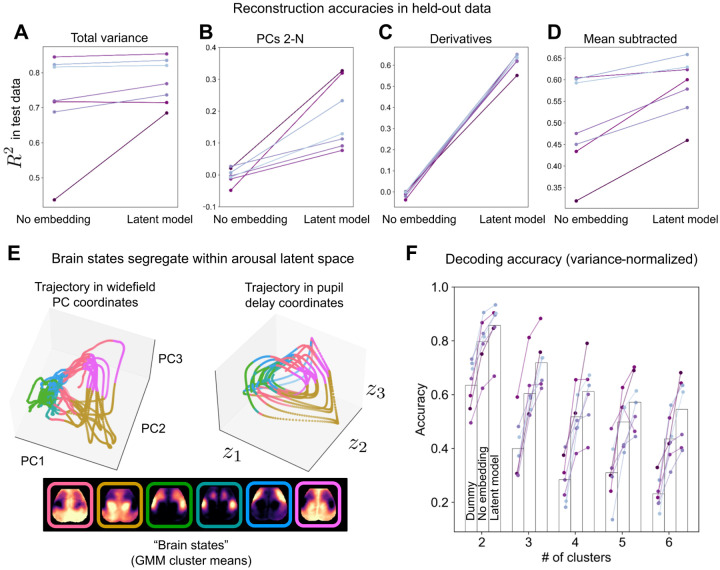
Figure 2: Pupil diameter enables reconstruction of spatiotemporal brain dynamics.
A Variance explained in widefield data test set (the last 3.5 minutes of imaging per mouse not used for model training). A simple linear regression model based on one (lag-optimized) pupil regressor (“No embedding”) accurately predicted 70–90% of variance in six of seven mice. Delay embedding and nonlinearities (“Latent model”) extended this level of performance to all mice, while further augmenting model performance, primarily by accounting for variance spread along higher dimensions (PCs 2-N) (B). C The latent model additionally accounted for the majority of variance in the temporal derivatives of widefield images in all seven mice, indicating that the dynamics of widefield images largely reflect the dynamics of arousal. The scalar, lag-optimized pupil regressor (i.e., “No embedding”) did not track the temporal dynamics of widefield images. D Although subtraction of the mean timecourse removed spatially uninformative variance, the latent model continued to explain a large fraction of the remaining, spatially-informative variance. E Example state trajectories (held-out data) visualized in coordinates defined by the first three widefield PCs (left) or three latent variables obtained from the delay-embedded pupil time series. Trajectories are color-coded by cluster identity as determined by a Gaussian mixture model (GMM) applied to widefield image frames (k = 6 clusters). Cluster means are shown below these trajectory plots. Color segregation in pupil delay coordinates indicates spatially-specific information encoded in pupil dynamics. F Decoding “brain states” from pupil dynamics. Comparison of classifier accuracy in properly identifying (high-confidence) GMM cluster assignments for each image frame based on the most frequent assignment in the training set (“Dummy” classifier), the “No embedding” model, or the “Latent model”. Classifier performance is shown as a function of allowable clusters (e.g., allowing for four possible clusters, pupil dynamics enabled > 60% classification accuracy.