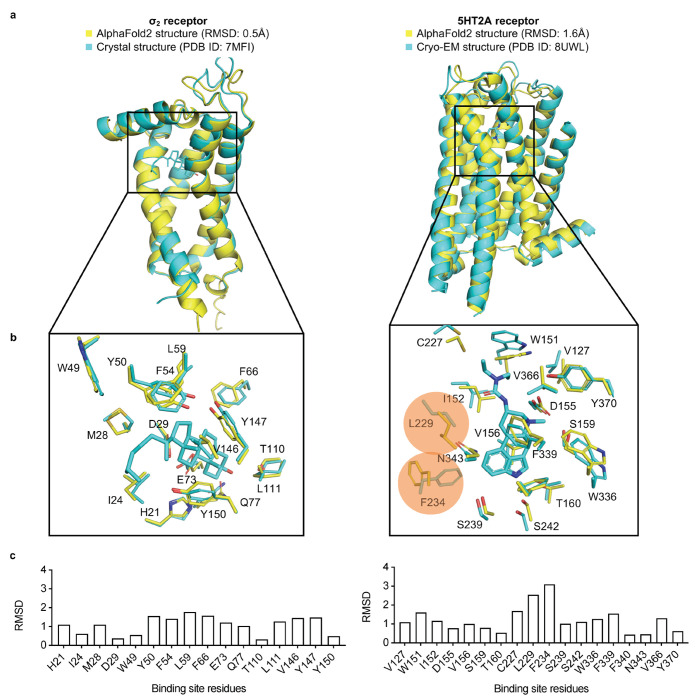
Figure 1 |. Structural comparisons of the σ2 receptor (left column) and the 5-HT2A receptor (right column) between the AlphaFold2 (AF2) predicted structure and the experimental structure.
a. The experimental structure (in cyan) is overlaid with the AF2 predicted structure (in yellow). The Root Mean Square Deviation (RMSD) value is calculated based on backbone atoms. The ligand binding site residues were selected within 5 Å distance from the ligand. b. The full-atom RMSD values of the binding site residues between the AF2 and the experimental structures. Two residues with large conformational differences between the AF2 and experimental structures used in docking, Leu229 and Phe234, are highlighted for the 5-HT2A receptor (right panel).