Figure 4. Egr1 overexpression partially recapitulates proestrus-associated changes in gene expression, chromatin, and synaptic plasticity.
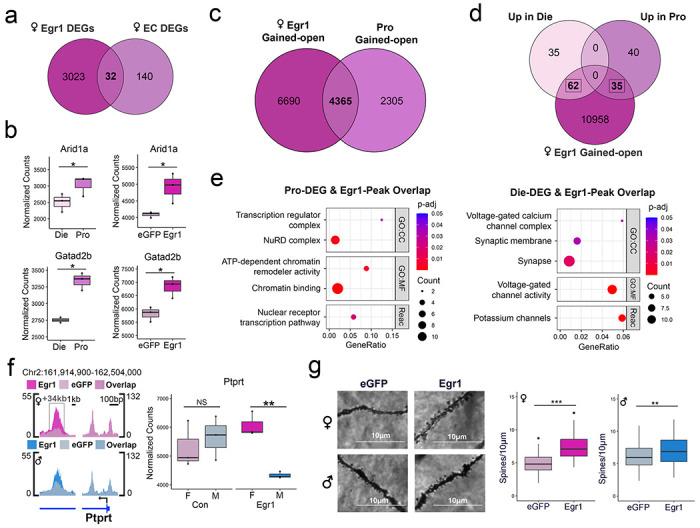
a. There is a significant overlap of Egr1 overexpression-induced and oestrous cycle (EC)-related DEGs. b. Genes upregulated in proestrus and induced by Egr1 overexpression include Arid1a and Gatad2b. c. There is a significant overlap of female Egr1-gained open chromatin regions and proestrus (Pro)-gained open chromatin regions. d. Oestrous cycle-related DEGs exhibit an even higher overlap with genes bearing Egr1 gained-open regions. e. The overlapping genes that are induced in proestrus and those that are downregulated in proestrus are enriched for different Gene Ontology (GO) terms. Dot plots colours indicate p-values and dot size corresponds to the number of overlapping genes between the list and the term/pathway (Count); the x-axis corresponds to the ratio of genes in the input list to genes in the term/pathway (GeneRatio). f. Ptprt is a synaptic function-related gene that shows sex-specific changes in chromatin accessibility and gene expression following Egr1 overexpression. ATAC-seq data is shown using SparK plots of group-average normalized ATAC-seq reads (n = 4 biological replicates/group/sex). RNA-seq data is shown using count plots (n=3 biological replicates/group/sex). Box plots (box, 1st-3rd quartile; horizontal line, median; whiskers, 1.5xIQR); RNA-seq significance codes; *, Padj<0.05; **, Padj<0.01; NS, non-significant; Benjamini-Hochberg correction for multiple testing. g. Egr1 overexpression increases dendritic spine density in the CA1 vHIP neurons of both OVX females and males, but the effect size is higher in females (d=1.532) compared to males (d=0.4468). Dendritic spine density significance codes; ***, P<0.001; **, P<0.01; Welch two-sample t-test. Die, dioestrus; Pro, proestrus; CC, Cellular Component; MF, Molecular Function; Reac, Reactome; F, female; M, male.
