Hidradenitis suppurativa (HS) is a human autoinflammatory disorder that primarily involves hair follicles in intertriginous skin areas of the groin, axillary, perineal, perianal, and inframammary regions (1). The clinical manifestations of HS are recurrent painful nodules and abscesses in sweaty areas of the body, fistulas, and severe scarring. With an early onset (in the early 20s), HS appears to have a female predominance (with a 3:1 sex ratio) and significant gender differences in epidemiology and disease manifestations (2). Although HS isn’t a contagious disease, it significantly compromises the quality of life caused by physical and psychological distress including intense pain, work disability, sexual dysfunction, social isolation, and depression (3). Unfortunately, HS is often associated with long diagnostic delays, due to disease unawareness among physicians. Further, HS has complex and heterogeneous histopathologic features and, in some cases, it is difficult to distinguish HS from other inflammatory skin diseases. The clinical features of HS appear to cover a broad histologic spectrum, which needs to be carefully considered. Smith et al. performed a comprehensive and systematic review of the histopathology of HS and confirmed a high prevalence of follicular occlusion, follicular hyperkeratosis, and hyperplasia of the follicular epithelium (4). Unfortunately, the medical treatment of HS is challenging due to the recurrent nature of this disease and the lack of detailed understanding of the pathogenesis of HS, which was originally thought to be an infectious condition of the apocrine sweat gland as suggested by its name (hidros meaning “sweat” and aden meaning “gland”) (3). In fact, there is no currently available curative therapy for HS. The existing treatments for HS are limited to only managing its symptoms, and surgical removal of affected skin areas remains the decisive option for nonresponder patients. Hence, HS presents an unmet medical need for which additional mechanistic understanding and therapeutic options are required. Based on current literature, inflammatory pathways have been considered the key factors in HS pathogenesis. Thus, the available therapies for the management of HS symptoms target various signaling molecules responsible for an exaggerated inflammatory response such as tumor necrosis factor alpha (TNF-α), and interleukins (IL) including IL-1β, IL-10, IL-17, and IL12/23 (5, 6). However, so far, the US Food and Drug Administration has only approved Adalimumab, a humanized monoclonal antibody against TNF-α, for the treatment of moderate-to-severe HS (5). Indeed, one of the main hurdles in the development of effective therapeutics is due to the lack of proper understanding of pathophysiological mechanisms of HS. Further, the interplay among genetics, hormonal status, inflammatory, and immune regulations remains unclear. The involvement of epigenetics in the pathophysiology of HS is not well understood. In addition, a detailed understanding of epithelial cell reprogramming in the context of chronic inflammation in HS is required. Moreover, studies focusing on immune dysregulation in HS are urgently needed to identify novel and specific pathways that may provide clues toward generating more effective treatments.
In this issue of PNAS, Jin et al. (7) present clinically relevant findings defining altered transcriptional states of the skin lesions at single-cell resolution in the human inflammatory disorder HS and determined the epigenetic reprogramming of basal stem/progenitor cells contributing to epithelial cell hyperproliferation and inflammatory pathogenesis. For this study, the authors conducted an elegant study with surgically discarded skin tissues from healthy and HS (age under 40, Hurley late II/III stage) patients. Using single-cell transcriptomics, they finely dissected the HS lesional skin and conducted multimodal molecular analysis. They performed single cell-RNA sequencing to characterize cellular heterogeneity, dual-omics profiling using CD49fhigh epithelial cells for chromatin accessibility and gene expression analysis, and epigenetic study using CUT&RUN (Cleavage Under Target & Release Using Nuclease) sequencing for multiple chromatin modification states.
In this issue of PNAS, Jin et al. present clinically relevant findings defining altered transcriptional states of the skin lesions at single-cell resolution in the human inflammatory disorder HS and determined the epigenetic reprogramming of basal stem/progenitor cells contributing to epithelial cell hyperproliferation and inflammatory pathogenesis.
Single-cell transcriptomics and spatial multiomics technologies are powerful tools that can be efficiently used to reveal hidden molecular complexities in disease pathogenesis (8) including HS (9–11). Using these cutting-edge methods, authors identified 19 highly distinct clusters of epidermal and immune cells in HS lesional skin. Further, using uniform manifold approximation and projection algorithm, they identified a HS-specific cluster within interfollicular epidermal cells, characterized by S100A inflammatory genes (S100A7/A8/A9). Subsequently, they determined a landscape of cluster-specific gene expression signatures for basal cell compartments and epidermal heterogeneity, which will be of high interest and a valuable reference for future studies focused on various human skin inflammatory diseases.
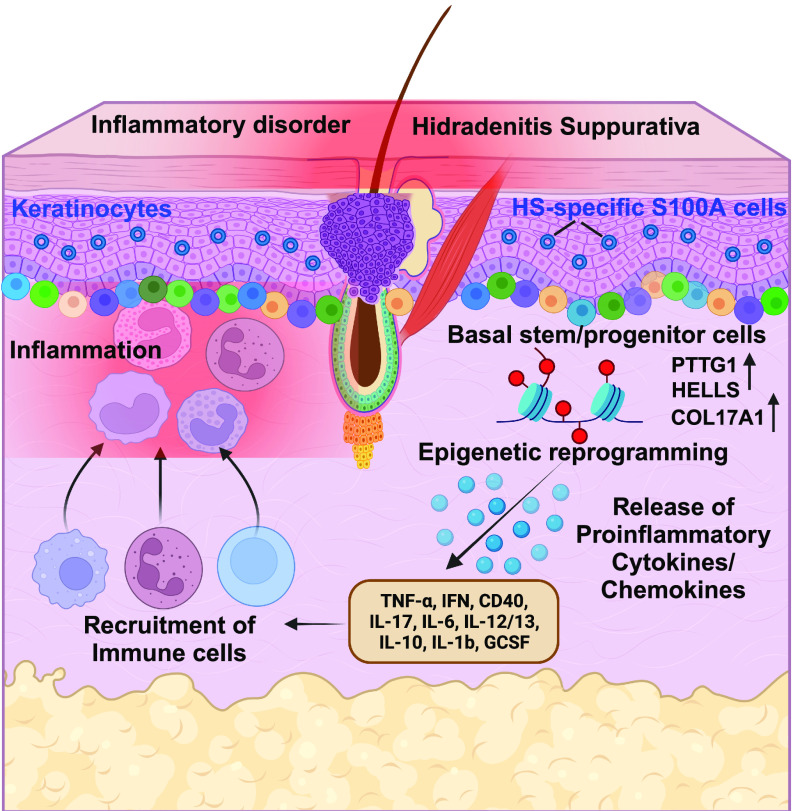
It is well known that inflammation is integral in HS pathogenesis, however, the contribution of the basal stem/progenitor cells to inflammation is unclear. Jin et al. (7) investigated to what extent the basal stem/progenitor cells redirect HS pathogenesis. Interestingly, they found twofold higher basal cells in HS patients than in healthy individuals. In addition, keratinocyte stem cells, characterized by CD49fbri/CD71dim staining (12), were found to be significantly higher in HS skin. Based on their observations, the authors suggest that the remodeling of the HS epidermis is spatially fueled by basal stem-like populations with niche-like compartments supporting the recruitment of immune cells (Fig. 1).
Fig. 1.
Epigenetic reprogramming in human skin inflammatory disorder HS. Compared to healthy skin, HS lesional epidermis is surrounded by heterogenous basal stem/progenitor cells with a significantly increased number of PTTG1+, HELLS+, and COL17A1+ cells favoring the recruitment of HS-associated immune cells. Stem cells lose their identity and differentiate to favor the formation of an epidermal cyst. HS-specific S100A+ keratinocyte cells amplify immune responses. After early inflammatory signals, substantial chromatin remodeling occurs in the genomic landscape of basal keratinocytes causing transcriptional activation of HS-associated genes to further drive recurrent inflammatory insults. The figure was created using Biorender.
At the molecular level, the increased expression of S100A family proteins in HS is known (13, 14); however, the physiological functions of these S100 proteins and their underlying mechanism in HS pathogenesis are largely unknown. To this end, Jin et al. (7) identified distinct signatures within the HS-specific S100A cells and demonstrated higher expression of proinflammatory response genes including S100A7/8/9, which suggests that these HS-S100A cells could be a new signaling center for aggravating the production of inflammatory cytokines (Fig. 1).
Epithelial stem cells have been shown to possess inflammatory memories—meaning they can remember an early inflammatory response and implement a more rapid response to subsequent stimuli (15). In this context, Jin et al. (7) found that the inflammatory environment in HS affected the chromatin landscape of keratinocyte stem cells using low-input ATAC and RNA-seq analyses of enriched CD49fhigh basal cells. Although these results are important in HS pathogenesis, it would be interesting for future studies if a similar concept could be applied to the inflammatory memory of epidermal stem cells, especially stem cells present in hair follicle (16), where HS begins.
Epigenetic mechanisms regulate gene transcription without changing the DNA sequence. So far, studies linking epigenetic mechanisms with skin inflammatory diseases (17) are mostly limited to atopic dermatitis and psoriasis (18, 19). A recent report linked epigenetic age acceleration with extrinsic immune-related changes in patients with HS (20). There is a clear knowledge gap in our understanding of the exact role of epigenetic mechanisms in HS pathogenesis. Jin et al. (7) investigated how epigenetic mechanisms could affect the transcription of HS-related genes by assessing the activity of the cis-regulatory elements employing CUT& RUN sequencing. Interestingly, they successfully identified several clinically relevant inflammatory enhancers and their coordinated transcription factors in HS basal CD49fhigh cells. Notably, this piece of data was also supported by in vitro experiments using CRISPR–Cas9-mediated genetic excision of one of the HS-specific enhancers in neonatal human epidermal keratinocytes. Similarly, the involvement of a transcription factor IRF3 (interferon-dependent immune response) displaying high expression across heterogenous populations of the HS epidermis was confirmed using a small-molecule inhibitor of IRF3 signaling BAY-985 (21), in HS keratinocytes.
Altogether, Jin et al. (7) have commendably integrated results from human clinical samples, single-cell sequencing of epidermal cells, primary basal keratinocytes culture, epigenetic mechanisms, and analysis of cytokine, chemokines, and inflammation mediators. This study is highly significant for characterizing the novel cellular and molecular signatures of HS epithelial cells and identifying the epigenetic mechanisms of basal progenitor cells leading to the inflammatory pathogenesis in HS. In addition, this study presents a novel database uncovering distinct chromatin accessibility patterns in epidermal progenitors in healthy and HS skin that could be a valuable reference for future studies in this area. Finally, these data pave a foundation for the development of future novel pharmacological agents targeting inflammatory enhancers and key transcription factors that could complement current biological drug-based therapies in HS.
Acknowledgments
N.A.’s research is supported by the National Cancer Institute of the NIH (R01 CA261937), and the Department of Veterans Affairs (Merit Review Awards; BLR&D I01BX005917; CSR&D I01CX002210; and a Senior Research Career Scientist Award BLR&D IK6BX006041). We would also like to acknowledge the support from the Dr. Frederic E. Mohs’s Skin Cancer Research Chair endowment to N.A.
Author contributions
G.C. and N.A. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
See companion article, “Epigenetic switch reshapes epithelial progenitor cell signatures and drives inflammatory pathogenesis in hidradenitis suppurativa,” 10.1073/pnas.2315096120.
References
- 1.Goldburg S. R., Strober B. E., Payette M. J., Hidradenitis suppurativa: Epidemiology, clinical presentation, and pathogenesis. J. Am. Acad. Dermatol. 82, 1045–1058 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Shih T., et al. , Gender differences in hidradenitis suppurativa characteristics: A retrospective cohort analysis. Int. J. Womens Dermatol. 7, 672–674 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinkel C., Thomsen S. F., Hidradenitis suppurativa: Causes, features, and current treatments. J. Clin. Aesthet. Dermatol. 11, 17–23 (2018). [PMC free article] [PubMed] [Google Scholar]
- 4.Smith S. D. B., Okoye G. A., Sokumbi O., Histopathology of hidradenitis suppurativa: A systematic review. Dermatopathol. (Basel). 9, 251–257 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldburg S. R., Strober B. E., Payette M. J., Hidradenitis suppurativa: Current and emerging treatments. J. Am. Acad. Dermatol. 82, 1061–1082 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Alikhan A., et al. , North American clinical management guidelines for hidradenitis suppurativa: A publication from the United States and Canadian Hidradenitis Suppurativa Foundations: Part II: Topical, intralesional, and systemic medical management. J. Am. Acad. Dermatol. 81, 91–101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin L., et al. , Epigenetic switch reshapes epithelial progenitor cell signatures and drives inflammatory pathogenesis in hidradenitis suppurativa. Proc. Natl. Acad. Sci. U.S.A. 120, e2315096120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandereyken K., Sifrim A., Thienpont B., Voet T., Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariottoni P., et al. , Single-cell rna sequencing reveals cellular and transcriptional changes associated with M1 macrophage polarization in hidradenitis suppurativa. Front. Med. (Lausanne) 8, 665873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J., et al. , Single-cell transcriptomics suggest distinct upstream drivers of IL-17A/F in hidradenitis versus psoriasis. J. Allergy Clin. Immunol. 152, 656–666 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Oliveira A., et al. , Transcriptome meta-analysis confirms the hidradenitis suppurativa pathogenic triad: Upregulated inflammation, altered epithelial organization, and dysregulated metabolic signaling. Biomolecules 12, 1371 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abreu C. M., Pirraco R. P., Reis R. L., Cerqueira M. T., Marques A. P., Interfollicular epidermal stem-like cells for the recreation of the hair follicle epithelial compartment. Stem Cell Res. Ther. 12, 62 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batycka-Baran A., Matusiak L., Nowicka-Suszko D., Szepietowski J. C., Baran W., Increased serum levels of S100A4 and S100A15 in individuals suffering from hidradenitis suppurativa. J. Clin. Med. 10, 5320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batycka-Baran A., et al. , Serum concentration and skin expression of S100A7 (Psoriasin) in patients suffering from hidradenitis suppurativa. Dermatology 237, 733–739 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Ordovas-Montanes J., et al. , Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng D., et al. , New insights into inflammatory memory of epidermal stem cells. Front. Immunol. 14, 1188559 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen B., Millar S., Skin epigenetics. Exp. Dermatol. 30, 1004–1008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt A. D., de Guzman Strong C., Current understanding of epigenetics in atopic dermatitis. Exp. Dermatol. 30, 1150–1155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng C., Tsoi L. C., Gudjonsson J. E., Dysregulated epigenetic modifications in psoriasis. Exp. Dermatol. 30, 1156–1166 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Lukac D., et al. , Increased epigenetic age acceleration in the hidradenitis suppurativa skin. Arch. Dermatol. Res. 315, 1037–1039 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Lefranc J., et al. , Discovery of BAY-985, a highly selective TBK1/IKKepsilon inhibitor. J. Med. Chem. 63, 601–612 (2020). [DOI] [PubMed] [Google Scholar]