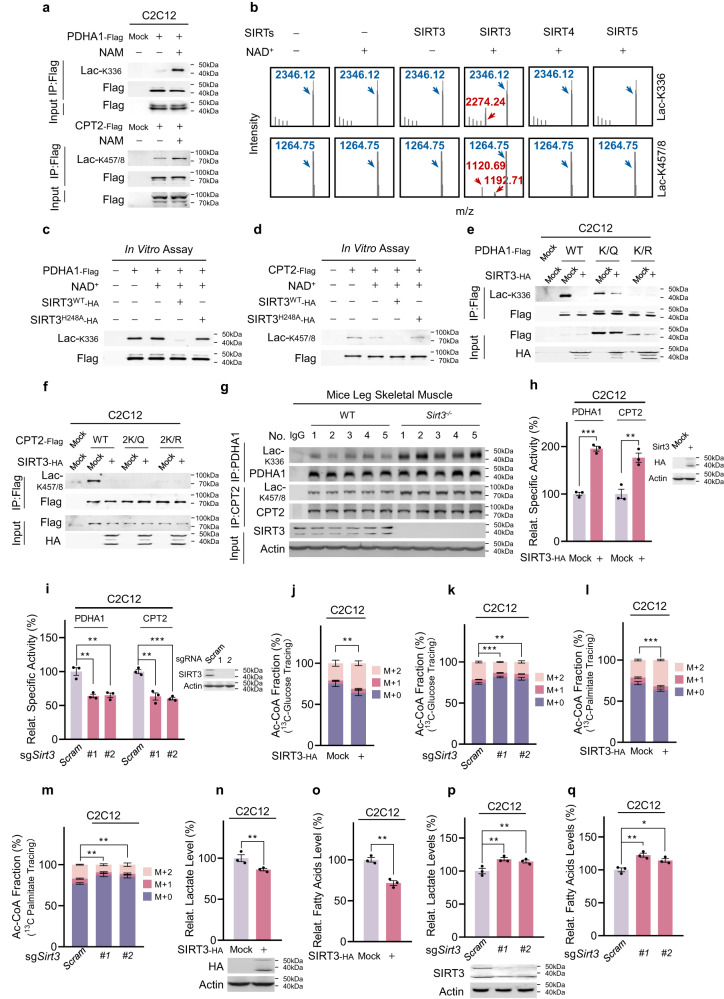
Fig. 6. SIRT3 reverses Lac-K336 and Lac-K457/8.
a NAM treatments increase Lac-K336 and Lac-K457/8. Lac-K336 and Lac-K457/8 levels of PDHA1 and CPT2 purified from C2C12 cells and NAM-treated C2C12 cells were detected. Cells were examined 3 h after 5 mM NAM treatment. b SIRT3 delactylates Lac-K336 and Lac-K457/8. The ability of SIRT3, SIRT4 catalytic domain and SIRT5 to delactylate synthetic Lac-K336- and Lac-K457/8-containing peptides was analyzed via MS to detect the formation of lactylated species. c–f SIRT3 decreases Lac-K336 and Lac-K457/8 levels. The Lac-K336 and Lac-K457/8 levels in purified PDHA1 (c) and CPT2 (d) were detected before and after being delactylated with SIRT3 or catalytic dead SIRT3H248A, and the SIRT3 expression effects on wild-type, lactylation-null PDHA1 (e), and CPT2 (f) were detected in C2C12 cells. g Ablation of Sirt3 increases Lac-K336 and Lac-K457/8 levels in vivo. Lac-K336 and Lac-K457/8 levels of wild-type and Sirt3−/− mouse leg skeletal muscles were detected (n = 5). h, i SIRT3 regulates PDHA1 and CPT2 activities. Relative activities of PDHA1 and CPT2 that were purified from C2C12 cells and SIRT3-overexpressing (h) or Sirt3 KO (i) C2C12 cells were compared (n = 3). j–m SIRT3 regulates Ac-CoA production from glycolysis and FAO. The M + 0, M + 1, and M + 2 fractions of Ac-CoA from 13C-glucose (j, k) and 13C-palmitate (l, m) were detected in C2C12 cells either overexpressing SIRT3 (j, l) or in which Sirt3 had been knocked out using independent sgRNAs (k, m) (n = 3). The chasing time for 10 mM 13C-glucose and 100 μM 13C-palmitate were 1 h and 12 h, respectively. n–q SIRT3 regulates lactate and free fatty acids levels. Lactate (n, p) and free fatty acid levels (o, q) in C2C12 cells overexpressing SIRT3 (n, o) or C2C12 cells in which Sirt3 had been knocked out via independent sgRNAs (p, q) were detected (n = 3). All data are reported as mean ± SEM of three independent experiments. Statistical significance was assessed by unpaired two-tailed Student’s t-test and two-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.001.