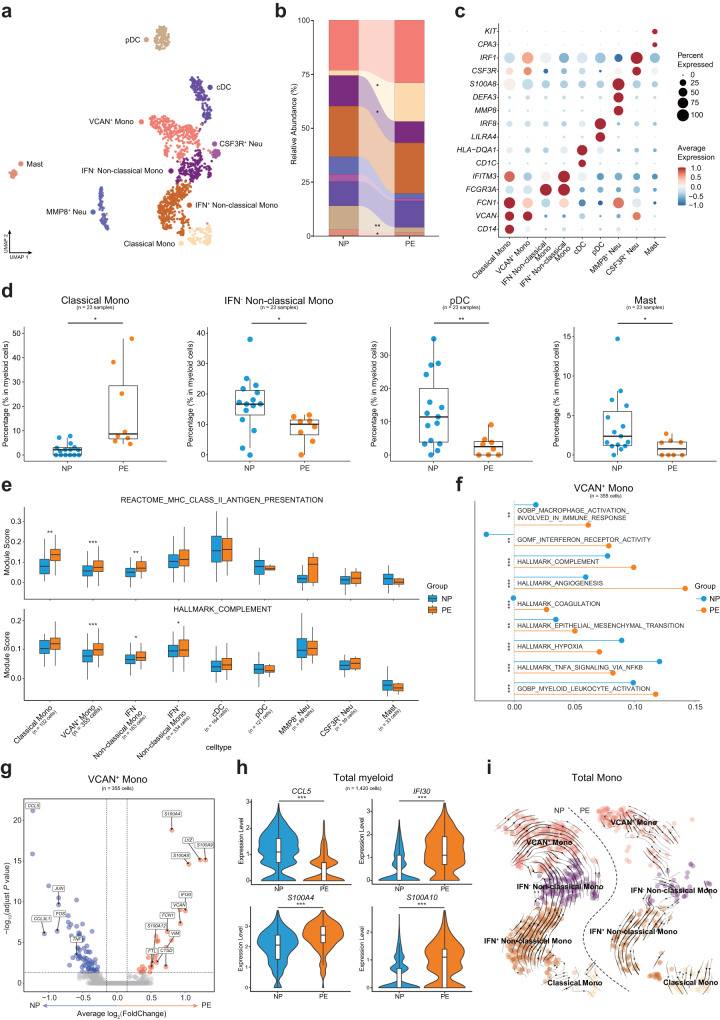
Fig. 4. Sub-clustering and analyzing the function of myeloid cell subsets.
a UMAP plot shows nine clusters of myeloid cell subsets of NP and PE. b Relative abundance of the nine clusters of myeloid cells in NP and PE. c Dot plot shows the expression of canonical markers which were used to identify myeloid cell subsets. d Box plots show the significantly changed percentages of myeloid cell subsets in total myeloid cells, Student’s t-test. *adjust P < 0.05, **adjust P < 0.01. e Box plots show the expression level of functional pathways in all myeloid cell subsets, Student’s t-test. **adjust P < 0.01, ***adjust P < 0.001. f Line and dot plot shows the expression of functional pathways in VCAN+ Mono, Student’s t-test. **adjust P < 0.01, ***adjust P < 0.001. g Volcano plot illustrates the significantly downregulated (blue dots) and significantly upregulated (red dots) DEGs in VACN+ Mono when comparing PE to NP, Wilcoxon rank-sum test. h Violin plots show the expression of functional genes in total myeloid cells, Wilcoxon rank-sum test. ***adjust P < 0.001. i UMAP plots show the developmental trajectories of all monocyte subsets in NP (left panel) and PE (right panel), the arrows indicate the differentiate directions. See also Supplementary Fig. 4 and Supplementary Data 3.