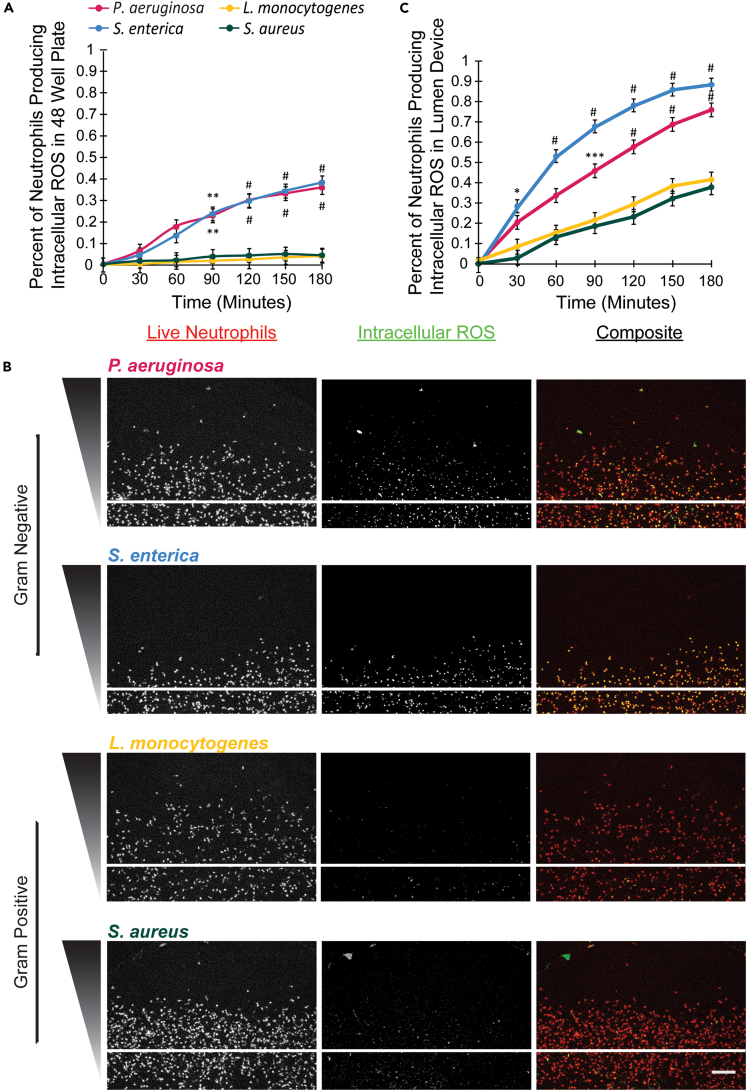
Figure 4.
An increased percentage of neutrophils produce ROS in response to gram-negative bacteria, P. aeruginosa and S. enterica
(A) Neutrophils were seeded in collagen gels in a 48-well plate in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus and stained with Calcein AM to visualize all live cells and DHR123 to visualize intracellular ROS production. The percentage of neutrophils producing intracellular ROS in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus was quantified by dividing the number of DHR123-positive neutrophils by the total number of neutrophils (Calcein AM). Data quantified from 3-well plates for each bacterial species across 3 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to both the L. monocytogenes and S. aureus condition. P values are labeled as ∗∗p <0 .01; #p <0 .0001.
(B) Neutrophils were seeded in the infection-on-a-chip device and stained with DHR123 to visualize intracellular ROS production following extravasation in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Representative images showing intracellular ROS production (DHR123) and total neutrophils (Calcein AM) in the infection-on-a-chip device in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Images were taken every 4 min for 8 h. Images shown are at 3 h after introduction of bacteria. The first column shows all cells stained red with Calcein AM, the second column shows DHR123-positive green, fluorescent ROS producing cells. Scale bar is 100 μm.
(C) The percentage of neutrophil expressing ROS was quantified in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Data quantified from 9 lumens for each bacterial species across 3 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to both the L. monocytogenes and S. aureus condition. P values are labeled as ∗p < 0.05; ∗∗∗p <0 .001; #p <0 .0001.