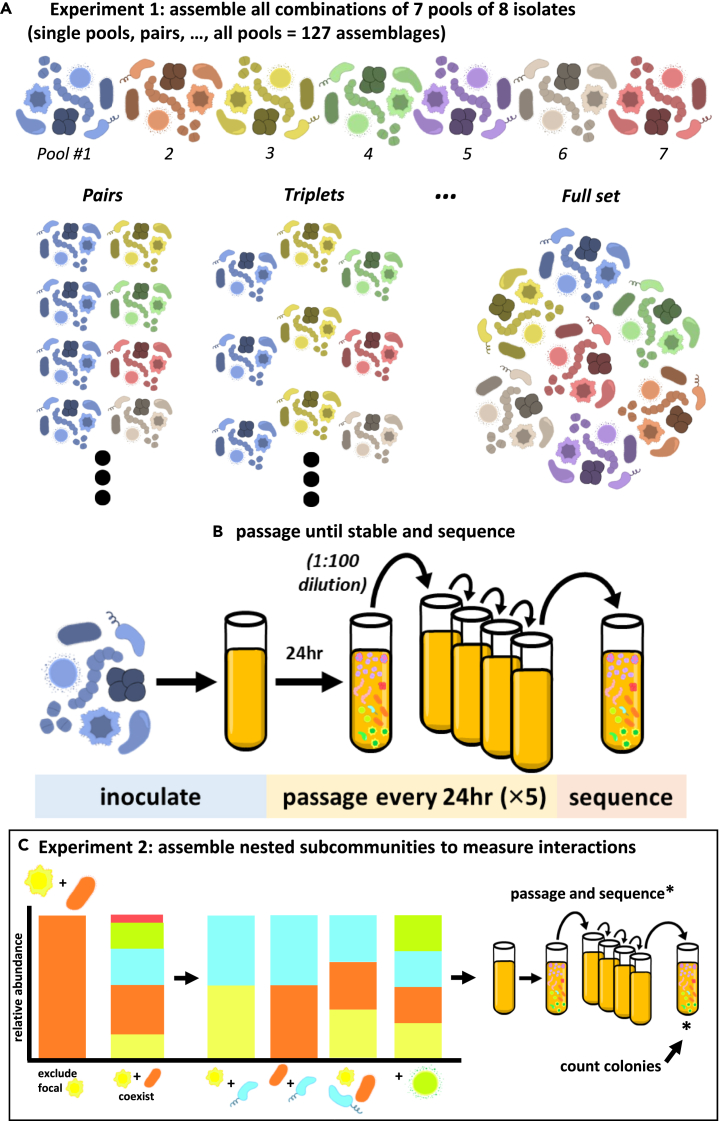
Figure 1.
Experimental outline
(A) A set of 56 isolates representing 21 genera were randomly pooled into 7 pools. All combinations of those pools were assembled at equal titers, with respective densities scaled to the total number of isolates initially present.
(B) These combinations were inoculated in triplicate into a custom medium derived from Arabidopsis leaves (ALM) and passaged daily into fresh medium at a 1:100 dilution for 5 days. To characterize the community compositions, the day-6 samples were sequenced, and short reads were mapped to reference genomes.
(C) Ten communities displaying context-dependent coexistence were decomposed into nested subcommunities containing the focal isolate and/or putative excluder isolate. These communities were assembled, passaged, and sequenced as described for the previous communities. To provide the absolute abundance information necessary to measure interactions, the final timepoint (day 6) was quantified by counting colonies on 1× TSA plates.