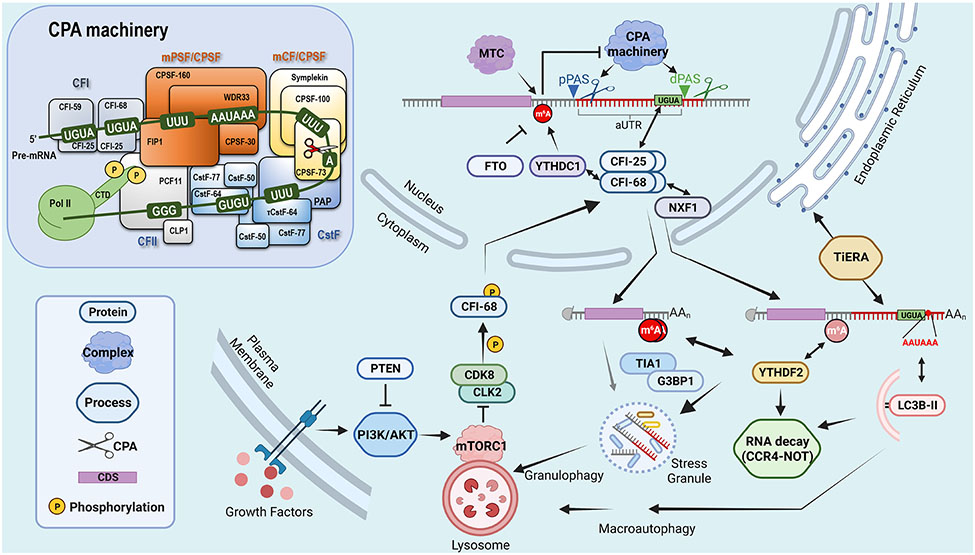
Figure 1.
Regulation of alternative polyadenylation (APA) site choice and differential mRNA metabolism between 3’UTR isoforms. A hypothetical gene with two 3’UTR APA sites are shown.
pPAS, proximal PAS; dPAS, distal PAS. MTC, m6A methyltransferase complex; LC3B-II, lipidated LC3B. CCR4-NOT carries out deadenylation of mRNA, often the first step for mRNA degradation. TiERA is translation-independent ER association. The core factors of CPA machinery and PAS motifs are also shown in the inset. Four subcomplexes in the machinery and two functional modules within CPSF are noted. CPSF-73 is the endonuclease responsible for precursor RNA cleavage, WDR33 and CPSF-30 collectively interact with the A[A/U]UAAA motif, and FIP1 binds to U-rich binding sequences. CstF exists as a dimer, each containing CstF-50, CstF-64/CstF-64τ and CstF-77. CstF-64 and CstF-64τ, two paralogs in the genome, have binding affinities to U-rich and GU-rich (GUGU or UGUG) motifs. CFI exists as a tetramer, comprising two molecules of CFI-25 together with two alternative larger subunits, While both CFI-59 and CFI-68 are RNA-binding proteins, CFI-25 has binding specificity to the UGUA motif. CFII includes CLP1 and PCF11, with the latter having binding affinity to G-rich RNA sequences 90.