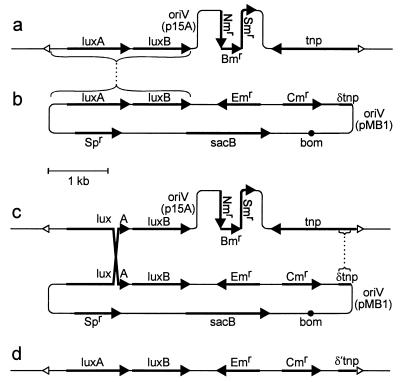
FIG. 1.
Marker exchange. (a) Transposon Tn5-1063 (45), bounded by open triangles, inserted in the genome (thin lines extending to the left and right) of Anabaena sp. (b) pRL386a, which contains genes luxA and luxB for recombination with inserted transposon Tn5-1063 and for transcriptional reporting. Shown are an erythromycin resistance gene (Emr) and a gene (Spr) conferring resistance to spectinomycin and streptomycin for selection of recombinants in Anabaena sp., with Spr also providing a simple screen to distinguish loss of the Spr-sacB portion from mutation of sacB; a chloramphenicol resistance gene (Cmr) for selection in E. coli; a portion (δtnp) of the transposase gene (tnp) to provide a second site for homologous recombination; a sacB gene to provide positive selection for double reciprocal recombination (8); an oriV (from plasmid pMB1) to allow replication in E. coli; and a bom site to allow conjugal mobilization. pRL386a is a derivative of pRL277 (7), which has unique BglII, dam-unmethylated XbaI, and NruI sites. V. fischeri luxAB as a BamHI fragment from pRL1063a was placed in the BglII site, cassette C.CE2 from pRL598a (6) was inserted in the dam-unmethylated XbaI site, and a 199-bp BalI-RsaI fragment from IS50R was transferred first to the EcoRV site of positive selection vector pRL487 and then moved as an NruI fragment to the pRL277-derived NruI site. The dotted lines link regions of sequence identity (brackets) that provide loci for homologous recombination. (c) Product of a recombinational event, obtained upon selection for resistance to erythromycin. (d) Product of a second recombinational event, obtained upon subsequent selection for resistance to erythromycin and sucrose. δ′tnp extends to the end of tnp.